Pharmaceutical Research for Digital Age Medicine
DSMZ scientist develops innovative screening method for medical agents
Advertisement
A team of scientists led by Professor Dr. Yvonne Mast, head of the Department of Bioresources for Bioeconomy and Health Research at the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures in Braunschweig, Lower Saxony, has developed a new screening strategy for active medical substances. This strategy is based exclusively on analyzing genome sequence data.

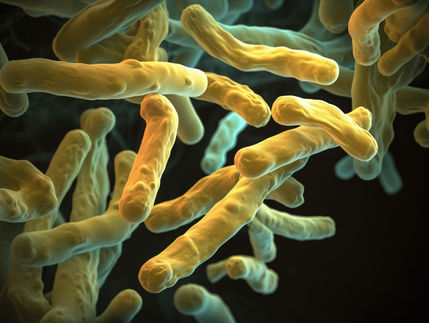
Symbolic representation of the Ψ-footprinting screening strategy for protein biosynthesis inhibitors. Shown are various actinomycetes on agar plates with a genome sequence (left), and the protein biosynthesis inhibitor amicoumacin inside a magnifying glass (right).
DSMZ/Mast

Prof. Dr. Yvonne Mast
DSMZ

Summarizing the advantages of her approach, Mast explains, "The advantage of our prediction strategy is that it can be used to screen purely digitally as well as very efficiently for specific potential drug producers and, in some cases, to screen out bacterial strains that are most likely to produce known substances. This makes the process of analyzing and finding active ingredients much easier. This new methodology is another important contribution in the rapidly developing research field of genome mining for the discovery of new active ingredients." In recent years, fewer and fewer new active ingredients, such as antibiotics, have been discovered and developed for medical use, while simultaneously the threat of multidrug-resistant pathogens have been rapidly increasing worldwide. A major problem in medical drug research is the high recovery rate of already known substances, which makes conventional screening methods highly inefficient.
Digitization in the field of active ingredient research
Digital sequence information is gaining more and more importance in active ingredient research. The team led by microbiologist Prof. Dr. Yvonne Mast has developed a prediction strategy based on genome sequence information from bacterial active ingredient producers that makes it possible to predict the production of a specific group of active ingredients, the so-called protein synthesis inhibitors. These active ingredients target the bacterial ribosome, which is an important place of action for many important antibiotics like erythromycin, tetracycline and clindamycin. The Braunschweig-based team of scientists were able to derive their prediction strategy by identifying specific indicator genes in the genome of drug producers from the bacterial group of Actinomycetes, which indicates protein biosynthesis inhibitor synthesis performance. The scientists provided proof of function by identifying the protein biosynthesis inhibitor amicoumacin from various Actinomyces strains of the DSMZ bacterial collection.