The ‘selenosome’: The choreography which governs recoding
Cryo-electron microscopy visualizes cellular structures
A team led by researchers from Charité – Universitätsmedizin Berlin has succeeded in deciphering a fundamental process of molecular biology: the incorporation of selenocysteine – the ‘21st amino acid’ – into selenium-containing proteins known as selenoproteins. These protein constructions are essential for the survival of humans and other mammals, as well as some microorganisms. However, the precise manner in which these proteins are synthesized and assembled in our bodies has remained unknown until now. Writing in Science, the authors provide the first detailed description of how a specific binding pocket interacts with other factors to facilitate this process.
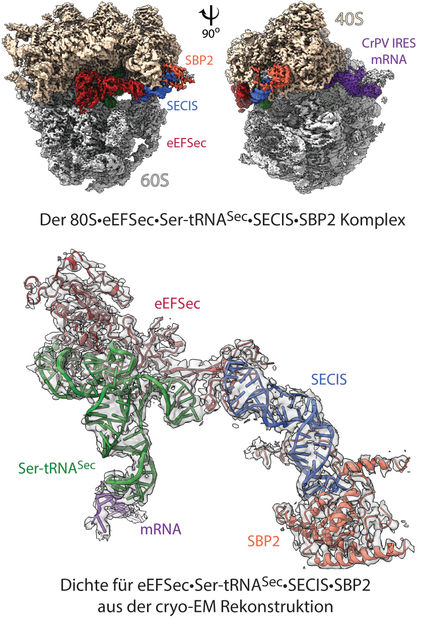
Structure of the ‘selenosome’: cryo 3D electron microscopy reconstruction of the ribosomal complex at resolution of 2.8 Å (top). The image shows the ribosome’s 40S and 60S subunits, with labeling indicating the binding elements (ligands): mRNA and other contributing factors. In the model (below), the highlighted density for the factors eEFSec•Ser-tRNASec•SECIS•SBP2 shows how the individual elements are arranged within the 80S ribosome, and how recoding is achieved.
© Charité | C. M. T. Spahn
Selenoproteins are an unusual group of proteins which were only discovered relatively recently. Scientists estimate there are up to 50 of these proteins in existence, yet only some of them have been studied. All selenoproteins have complex structures which center around at least part of the amino acid which gives them their name: selenocysteine (Sec). Selenoproteins play an important protective and defensive role in the human body, both at the cellular and organism level. They primarily function as oxidoreductases, i.e., mediators of key chemical reactions, and as mediators of thyroid function. Selenoproteins are also thought to help protect the body against cancer, as they contain the element selenium, which is capable of quickly reducing oxidative stress.
How are these special proteins formed? What is the molecular choreography which sets out the steps involved in the incorporation of selenocysteine during protein biosynthesis? What is the precise structure of the selenosome, the complex which forms to facilitate the synthesis of selenoproteins? Working alongside partners at the Max Planck Institute for Molecular Genetics, the University of Illinois Chicago, and the Rutgers Robert Wood Johnson Medical School in New Jersey, the team led by Prof. Dr. Christian Spahn, Director of the Institute of Medical Physics and Biophysics at Charité, and Dr. Tarek Hilal of the Institute of Chemistry and Biochemistry at Freie Universität Berlin, used three-dimensional, high-resolution cryo-electron microscopy to visualize and understand the structures and steps involved in this fundamental molecular biology process.
Ribosomes, known as the protein factories of the cell, usually synthesize proteins by following strict blueprints delivered by messenger RNA (mRNA) molecules. The universal genetic code uses specific sequences consisting of triplet combinations (codons) of four different bases. Each codon forms a unit of genetic information encoding a particular amino acid. However, the specific structure of selenoproteins requires more than the basic set of 20 amino acids. All selenoproteins contain selenocysteine (Sec), which is known as the 21st amino acid.
Selenocysteine does not have its own designated codon, meaning that, unlike the other amino acids, it is not encoded by a specific base triplet. Ribosomes must therefore acquire a very special set of features during selenoprotein synthesis. The ribosome is reprogrammed by a special mRNA signaling sequence known as the SECIS element (selenocysteine insertion sequence), which effectively overwrites the existing genetic code. A specific codon – which normally signals the termination of the translation process and hence the release of the emerging protein chain (UGA stop codon) – is thus recoded to insert selenocysteine into proteins. In addition to the SECIS element, this recoding process also requires a specific transfer RNA species (tRNASec) which carries selenocysteine and other special translation factors. “While the players involved have been known for several years, their precise functions and interactions had remained a mystery until now,” says Prof. Spahn, adding: “A particular mystery was the precise action of the SECIS element, which is not positioned in close proximity to the recoded UGA stop codon, but at the very end of the mRNA linear sequence, many hundreds of nucleotide building blocks away.”
In order to elucidate the molecular mechanism involved, the researchers started by recreating the ‘selenosome’, the ribosomal complex which forms in order to recode the UGA stop codon, in the laboratory. Using a high-resolution imaging technology known as cryo-electron microscopy, the researchers then produced a 3D visualization of the tiny complex, which enabled them to study its structure. “Using our structural visualizations, we were then able to determine how the factors involved interact with the ribosome and how these interactions reprogram the ribosome,” explains Prof. Spahn. “Specifically, we were able to show that the mRNA forms a large loop, meaning that the UGA stop codon and the SECIS element bind to the ribosome at the same time. The SECIS element is anchored via a previously unknown binding pocket on the ribosome and, while bound to the ribosome, can promote the insertion of selenocysteine with the assistance of translation factors.” Both the selenosome’s structure and its mechanism came as a surprise to the researchers. Neither could have been predicted, because the process involved in selenocysteine insertion in humans and other mammals is distinctly different from that known to take place in bacteria. The complex described in this study is an example of the way in which signaling structures at the back end of the mRNA strand (i.e., outside the coding region) can interact with the ribosome in order to control its function.
The researchers were able to elucidate the very first step in the selenocysteine insertion process. Subsequent steps remain unclear and will be addressed in future studies. Research such as this helps to increase our understanding of the vital function of the trace element selenium in the body, as well as the significance of its role in normal physiology and the development of diseases like diabetes and cancer.