Design of protein binders from target structure alone
A new method for generating potent, specific binding proteins yields candidate medicines for cancer, diabetes, inflammation and more
A team of scientists has created a powerful new method for generating protein drugs. Using computers, they designed molecules that can target important proteins in the body, such as the insulin receptor, as well as vulnerable proteins on the surface of viruses. This solves a long-standing challenge in drug development and may lead to new treatments for cancer, diabetes, infection, inflammation, and beyond.
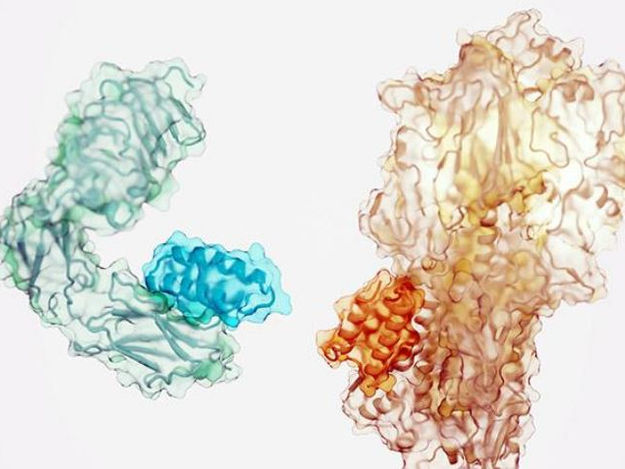
Small proteins (darker shade) designed to bind to the insulin receptor (left) and a component of the influenza virus (right).
ian Haydon/Institute for Protein Design
The research, appearing in Nature, was led by scientists in the laboratory of David Baker, professor of biochemistry at the University of Washington School of Medicine and a recipient of the 2021 Breakthrough Prize in Life Sciences.
“The ability to generate new proteins that bind tightly and specifically to any molecular target that you want is a paradigm shift in drug development and molecular biology more broadly,” said Baker.
Antibodies are today’s most common protein-based drugs. They typically function by binding to a specific molecular target, which then becomes either activated or deactivated. Antibodies can treat a wide range of health disorders, including COVID-19 and cancer, but generating new ones is challenging. Antibodies can also be costly to manufacture.
A team led by two postdoctoral scholars in the Baker lab, Longxing Cao and Brian Coventry, combined recent advances in the field of computational protein design to arrive at a strategy for creating new proteins that bind molecular targets in a manner similar to antibodies. They developed software that can scan a target molecule, identify potential binding sites, generate proteins targeting those sites, and then screen from millions of candidate binding proteins to identify those most likely to function.
The team used the new software to generate high-affinity binding proteins against 12 distinct molecular targets. These targets include important cellular receptors such as TrkA, EGFR, Tie2, and the insulin receptor, as well proteins on the surface of the influenza virus and SARS-CoV-2 (the virus that causes COVID-19).
“When it comes to creating new drugs, there are easy targets and there are hard targets,” said Cao, who is now an assistant professor at Westlake University. “In this paper, we show that even very hard targets are amenable to this approach. We were able to make binding proteins to some targets that had no known binding partners or antibodies,”
In total, the team produced over half a million candidate binding proteins for the 12 selected molecular targets. Data collected on this large pool of candidate binding proteins was used to improve the overall method.
“We look forward to seeing how these molecules might be used in a clinical context, and more importantly how this new method of designing protein drugs might lead to even more promising compounds in the future,” said Coventry.
Most read news
Other news from the department science

Get the life science industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.
Most read news
More news from our other portals
Last viewed contents
Mange
Fletcher's_Laxative
Manzanita
Li_Ching-Yuen
Cannabis_(drug)
Hiatus_hernia
Sadistic_personality_disorder
Rigidity
BTI researcher gets NSF grant to create mutant maize lines for research