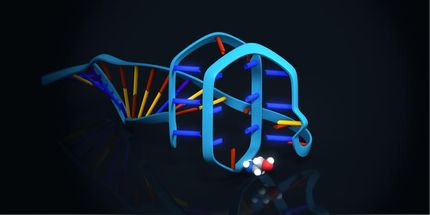
Novel hepatitis C virus inhibitory molecules by in vitro evolution
This work could contribute to the development of new biosensors and diagnostic systems for hepatitis C
An interdisciplinary CSIC team, formed by researchers from the Center for Astrobiology (CAB, CSIC-INTA), the Center for Molecular Biology Severo Ochoa (CBMSO, CSIC-UAM) and the National Center for Biotechnology (CNB, CSIC), has obtained short DNA and structured RNA molecules that inhibit hepatitis C virus (HCV) replication in cell culture.
Recreation of the hepatitis C virus.
Bruce Blaus-Wikimedia
These molecules, known generically as "aptamers", have been produced and characterized at the CAB using variants of a system called SELEX. These aptamers, already patented, bind with high affinity and specificity to the protein known as "core", which forms the HCV capsid and is the most conserved among those encoded by its genome.
In the words of Carlos Briones, CAB researcher and lead author of the work: "it is very interesting to see how the technology of in vitro molecular evolution of nucleic acids, which we use in research on the origin of life and the RNA World, also allows us to obtain useful molecules in the fields of diagnosis and therapy, both for HCV and against other viruses with which we work".
Bioinformatics analysis of the aptamers obtained, carried out mainly at the CNB, has identified the sequence motifs present in the molecules that bind to the core protein of HCV genotypes 1 to 4, which are responsible for 95% of cases worldwide (about 4 million new infections per year). Furthermore, cell culture assays performed at the CBMSO have shown that these aptamers do not present toxicity for the cell, and that at least two of them have a high capacity to inhibit virus replication, interfering with the formation of its capsid.
This work, published in the Journal of Molecular Biology, could contribute to the development of novel biosensors and diagnostic systems for hepatitis C, with a much higher detection sensitivity than the current ones. Furthermore, according to Esteban Domingo, co-author of the study, "these aptamers could give rise to novel therapeutic options against a virus as prevalent worldwide as HCV, becoming alternative or complementary antiviral drugs to the current ones, which are very expensive and against which cases of resistance have already been detected". Such applications in diagnosis and therapy are currently being explored in collaboration with different Spanish and foreign research groups.
Three of the authors of this work (Celia Perales, Esteban Domingo and Carlos Briones) also belong to the Centro de Investigación Biomédica en Red de enfermedades hepáticas y digestivas (CIBERehd), of the Instituto de Salud Carlos III. Moreover, this research is in line with the interests of the LifeHUB.CSIC initiative, recently created at CSIC to promote studies on the origin and early evolution of life.
Note: This article has been translated using a computer system without human intervention. LUMITOS offers these automatic translations to present a wider range of current news. Since this article has been translated with automatic translation, it is possible that it contains errors in vocabulary, syntax or grammar. The original article in Spanish can be found here.
Original publication
B. Torres-Vázquez, A.M. de Lucas, C. García-Crespo, J.A. García-Martín, A. Fragoso, M. Fernández-Algar, C. Perales, E. Domingo, M. Moreno, C. Briones; "In vitro selection of high affinity DNA and RNA aptamers that detect hepatitis C virus core protein of genotypes 1 to 4 and inhibit virus production in cell culture."; Journal of Molecular Biology 2022.