Unmuting the genome
Epigenetic mechanisms for parent-specific genetic activation decoded
hereditary diseases as well as cancers and cardiovascular diseases may be associated with a phenomenon known as genomic imprinting, in which only the maternally or paternally inherited gene is active. An international research team involving scientists at the Technical University of Munich (TUM), the Max Planck Institute for Molecular Genetics (MPIMG) in Berlin and Harvard University in Cambridge (USA) has now investigated the mechanisms responsible for the deactivation of the genes.
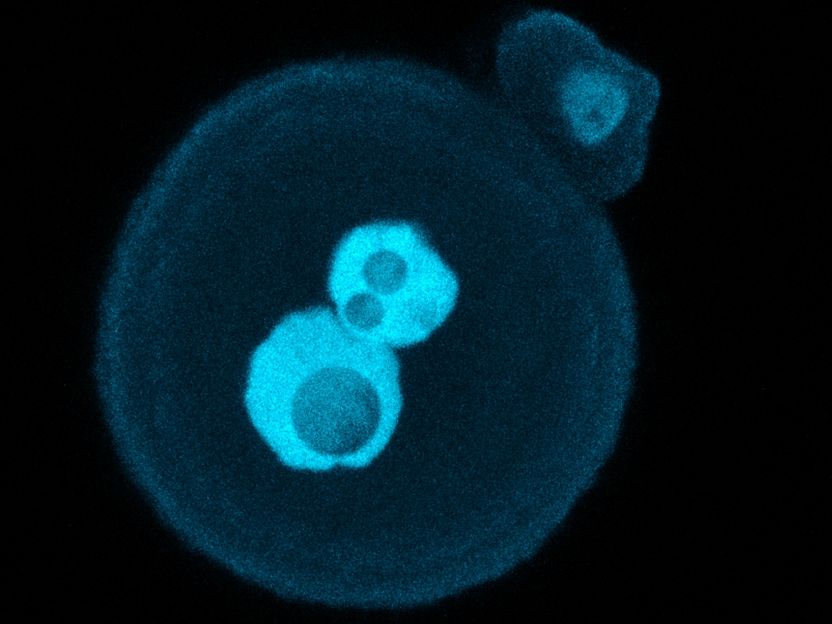
The zygote of a mouse stained with fluorescent dyes, with the nuclei of the sperm and egg cells still visible as bright blobs in the middle
A. S. Kumar MPI f. mol. Genet.
Our cells contain the entire genetic information from our mother and our father. From each of them we inherit 23 chromosomes that contain our DNA. Two copies of each gene are therefore present in our genome and, as a general rule, both are active. This has the advantage that defective mutations inherited from the mother or father are generally cancelled out by the other copy of the gene.
However, for around one percent of our genes, only the gene inherited from the father or mother is active, while the other is deactivated, a phenomenon known as genomic imprinting.
Approach for treating diseases
“Many genetic and epigenetic diseases are associated with genomic imprinting, such as Beckwith-Wiedemann syndrome, Angelman syndrome and Prader-Willi syndrome,” explains Daniel Andergassen, the head of the Independent Junior Research Group at the Institute of Pharmacology and Toxicology at TUM. “If the healthy, deactivated gene could be reactivated, it would be theoretically possible to compensate for complicationscaused by the active, defective gene.”
“But before developing future treatments, we need to understand the fundamentals,” says Alexander Meissner, director at the MPIMG. “It has become clear in recent years that genomic imprinting is mediated by multiple molecular mechanisms.”
Read lock for the gene
In genomic imprinting, either the “packaging” of the genetic material or the DNA itself is chemically modified. Instead of the genetic information being changed, the modifications block the gene from being read.
“These are so-called epigenetic mechanisms,” says Andergassen. “The DNA can be seen as the hardware, and epigenetics as the software responsible for regulating the genes.” Genetic regulation takes place in every cell in the body. All the cells contain the same genetic information, but depending on the organ, different genes are active.
Genetic scissors remove the “off switch”
Meissner and Andergassen, who at the beginning of the study where still conducting the research at Harvard University (USA) along with Zachary Smith, used mice to investigate which epigenetic mechanisms were behind the imprinting.
They used the molecular biology technique known as CRISPR-Cas9 that functions as a “genetic scissors”, removing and inserting segments of DNA. The scientists removed known epigenetic “off switches” and observed whether the deactivated gene was reactivated. With this approach, they were able to link the most important epigenetic “off switches” with imprinted genes.
Hydrocarbon molecules render genes inactive
It turns out that most of the genes are inactivated through DNA methylation that attaches hydrocarbon molecules to the genetic material. Another group of genes is silenced by a set of enzymes known as Polycombs. In the placenta, an additional mechanism comes into play: In this tissue, some genes are deactivated by chemically modifying the proteins that serve as a structural scaffold for the DNA.
The small but crucial difference
Along with genomic imprinting that switches off individual genes, the researchers investigated another phenomenon. In female cells, which unlike male cells have two X chromosomes, one chromosome is entirely deactivated very early in embryonic development. This is true in almost all mammals, including humans.
“We discovered that the enzyme PRC2 plays an important role in the inactivation of the X chromosome, at least in the placenta,” says Andergassen. “Once we remove this enzyme, the silent X chromosome is reactivated.” The results could be significant for X-chromosome-related disease because reactivation of the silent gene could compensate for the malfunctioning active gene.
In a follow-up project at TUM, Andergassen will study whether heart diseases might also be associated with epigenetics and especially with the inactive X chromosome in women. “Because our epigenetics change as we get old, it is conceivable that the X chromosome becomes active again and that the duplicate genetic activity has a negative influence,” says the researcher.
With their research, the team succeeded in providing an overview of the epigenetic mechanisms that maintain genomic imprinting. “We can explain virtually all parent-specific gene expression with the three known epigenetic mechanisms,” says Andergassen. “However, we know relatively little about expression in the placenta and whether it is similar in all mammals. Further studies will be needed to investigate how these processes influence the development of the fetus.”