New microscopy method offers 3D tracking of 100 single molecules at once
Multipurpose technology
Since its invention over 400 years ago, the microscope has continued to evolve, peering ever deeper into nature’s mysteries at the smallest scales.

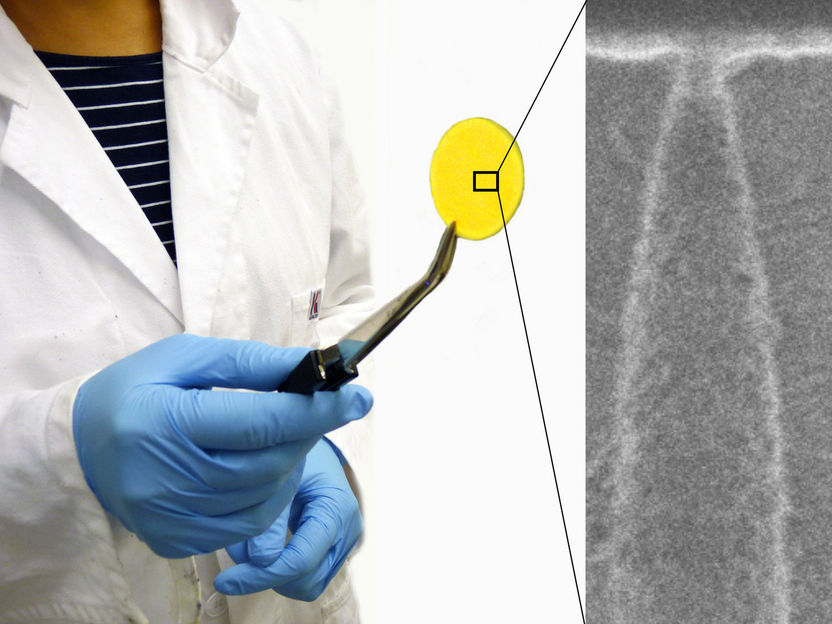
The graphic shows the basic experimental setup for Surface Plasmon Resonance Microscopy (SPRM). When a particle of interest binds with a receptor, tethered to a thin gold film, the event disturbs a surface plasmon wave, which is registered as a change in light intensity. SLED (for super-luminescent emitting diode) is the light source, which illuminates the sample and induces a surface plasmon wave that ripples across the gold surface. A high speed camera captures these rapid changes, imaging the binding dynamics.
Adapted with permission from Three-Dimensional Tracking of Tethered Particles for Probing Nanometer-Scale Single-Molecule Dynamics Using a Plasmonic Microscope Copyright 2021 American Chemical Society.
In new research, Prof. Shaopeng Wang, his postdoctoral research scholar Guangzhong Ma along with their colleagues at the Biodesign Center for Bioelectronics and Biosensors at Arizona State University describe advances in microscopy based on a phenomenon known as surface plasmon resonance (SPR).
The new study highlights a series of experiments that show how SPR technology can be used to precisely image 100 single molecules simultaneously and describe their dynamic activities in real time, particularly when molecules bind with each other and carry out important biological functions.
“Taking advantage of the extremely high sensitivity in the axial (vertical) direction, SPR can track the particle's axial motions with sub-nanometer precision, which is much more precise than regular microscopy imaging,” Wang says. “We demonstrated that this feature can be used to study the details of molecular binding events at the single molecule-level and also for processing multiple signals at once.”
New wave science
SPR is an optical effect that can be used to detect and precisely measure the binding of molecules without the need for fluorescent labels. The technique has broad applications, both in industry and the medical field, where it can be used to screen and develop new pharmaceutical drugs and biotherapeutics, new diagnostic assays, and unlock some of the mechanisms of disease.
Many critical biomolecules in health and disease are challenging to observe using conventional techniques such as X-ray crystallography or NMR spectroscopy. SPR offers an efficient and low-cost alternative for these investigations.
SPR relies on the fact that metals contain many electrons that are not bound to atoms. When incident light is shined on these free electrons under the proper conditions, the energy contained in the light causes these electrons to resonate, producing a wave across the metal surface, known as a surface plasmon.
The SPR technique can be used to measure the binding between proteins, nucleic acids, small molecules, and many other interactions. Precise explorations of such biomolecules are challenging as most measure just a few nanometers in size. The resulting data can help researchers identify which molecules in a sample interact, why they interact, and the strength of these interactions.
SPR is one of the only techniques that allows researchers to study not only the binding affinity of molecules but also their binding kinetics, yielding valuable information not captured by traditional tests such as ELISA. Surface Plasmon Resonance Microscopy (SPRM) takes the basic SPR phenomenon a step further, producing a highly versatile technology for investigating molecular binding events at sub-nanometer scales and with millisecond time resolution.
Sensitive detections
In order to use SPR to delicately probe the binding of a single molecules, the receptor molecule, such as a short segment of DNA, is immobilized on a sensor surface composed of a very thin layer of gold. Then the binding molecule is added to an aqueous solution.
When polarized light is directed onto the gold film at just the right angle, plasmonic waves are generated. A binding event of the immobilized receptor and the binding molecule can be detected when it alters the refractive index at the gold surface. This effect acts to disturb the surface plasmon, producing an increase in signal intensity. These fleeting effects are then captured with the aid of high-speed cameras.
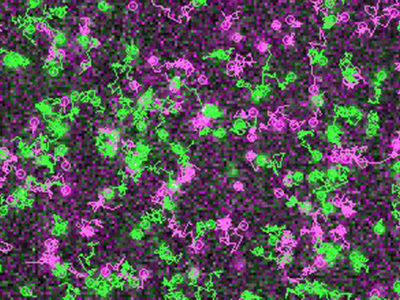
In the current study, SPR imaging was used to track 100 particles simultaneously in three dimensions. In one experiment, bound segments of double-stranded DNA, 48 base pairs in length and affixed to a gold surface, acted as the receptor molecules. The molecule they were poised to detect was a small enzyme known as a helicase. Its role in biological systems is to bind with and unwind DNA, when repairs to the DNA sequence are required.
The DNA helicase was first attached to a gold nanoparticle, to allow SPR to reveal its subtle optical properties as it bound with the tethered DNA strand.
With the addition of ATP to the aqueous solution, the helicase set about its task, unwinding the double-stranded DNA at a rate of roughly 10 base pairs per second, with the live action captured by a 400 fps camera. The high-precision 3D tracking method outlined in the study enabled the researchers to derive the unwinding rate of the DNA strand as well as the rotation angle of the bound helicase molecule.
Multipurpose technology
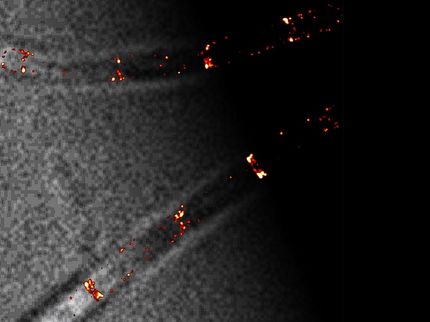
Other experiments demonstrate that the method can perform particle-based detection of antibody-antigen binding and discriminate between specific and non-specific binding events. (Non-specific antibody binding can occur when a cell lacks a receptor or epitope for a specific antibody.) Here, the molecule troponin T, a common biomarker for heart disease, was observed as it bound to its corresponding antibody affixed to the gold surface.
The ability to separate specific from non-specific binding events is a valuable benefit, as the latter often occur in mixed samples like serum, making accurate diagnostic analysis challenging.
Further, because the SPRM technique can capture data on 100 molecules at once in 3 dimensions, the method allows detailed statistical analysis of molecular binding events and kinetics not possible with conventional methods. Thus, the new technique shows considerable promise for rapid, low-cost, and detailed detection of biomolecules and may be incorporated into a new generation of biosensing devices.