Enzyme silences virus-like sequences
Methyltransferase Dnmt1 is not only responsible for maintenance
Advertisement
Often, biology will not fit into clear-cut categories. In a recent publication, an enzyme to which researchers had previously assigned a distinct task is stepping outside its box. It not only maintains the epigenetic state of the genome, but can also specifically silence additional DNA segments with virus-like characteristics.
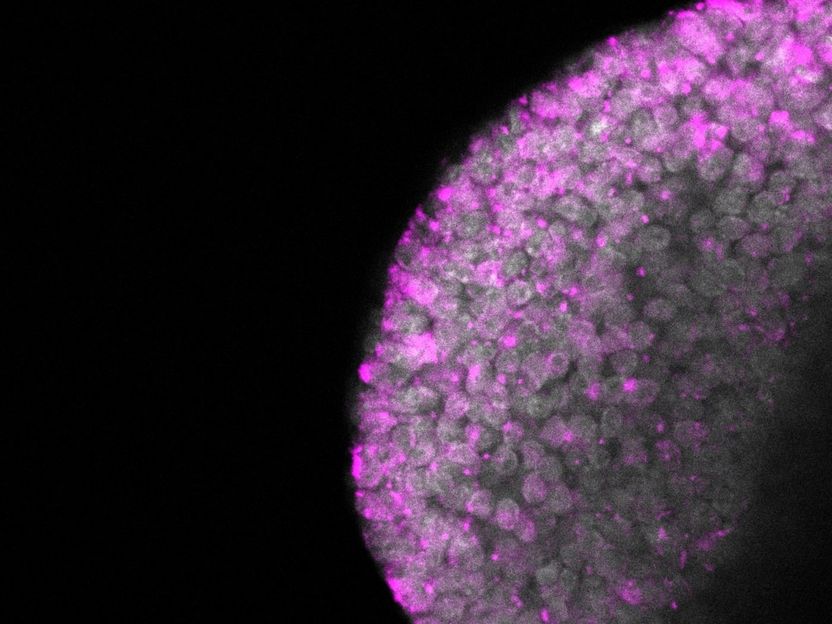
After the methyltransferase Dnmt1 is knocked out in a zygote, IAP retrotransposons are upregulated (retrotransposon molecules labeled pink)
Abhishek Sampath Kumar, MPI f. mol. Genet
It is the forbidden section in the library of life: Some parts of the DNA contain harmful information. Methyltransferase enzymes provide these sections with chemical labels, causing them to be ignored by the cell. Like the librarian putting warning labels on certain books, these enzymes leave the genetic sequence untouched by appending small carbon packets (methyl groups) to the DNA molecule, which means they are operating epigenetically.
For decades, there seemed to be a clear division of labor: the methyltransferases Dnmt3a and Dnmt3b hunt for suitable segments of DNA and garnish them with methyl groups. The sibling enzyme Dnmt1 then merely copies labels when only one side of the DNA strand is methylated after cell division or where one half of the two strand’s labels have been lost.
However, it is not as simple as that, as Alexander Meissner's research group at the Max Planck Institute for Molecular Genetics shows in a new paper published in Nature Structural and Molecular Biology. According to the new study, Dnmt1 has, contrary to previous assumptions, the ability to target specific sequences in the mouse genome and attach methyl marks to them outside of its maintenance duties.
The fertilized egg loses its methylation until the early embryo turns into a bubble-like state, the blastocyst. “The blastocyst has one of the lowest overall levels of DNA methylation during mouse development, but just 72 hours later, the genome has transitioned to a highly methylated state”, Meissner says. “This is why we took a closer look at these particularly interesting time points.”
But why is it necessary to turn off specific DNA segments in the first place? Cells use methylation to determine which genetic programs to run. Yet there is a more insidious reason, Meissner explains: “Most genomes – including those of humans and mice – contain thousands of virus-derived sequences that have embedded themselves in the genomes of their hosts over the last few million years.”
The virus-like sequences are transposons, also known as “jumping genes”. They have the ability to spontaneously replicate and integrate anywhere in the DNA. Transposons have spread throughout the genome and make up roughly 40 per cent of the genome in mice and humans.
“Methylation helps to keep these potentially harmful stretches of the genome in check,” says Chuck Haggerty, doctoral candidate and one of the publication's first authors. “If a transposon or virus jumps into the middle of a gene, it could alter its function. And if transposons were allowed to spread uncontrollably throughout the genome, its integrity could be at risk.”
“We noticed that the genome of embryos still gained methylation even when we knocked out Dnmt3a and 3b,” Haggerty says. The researchers modeled this in a cell culture, knocked out Dnmt3a and 3b again, and then additionally switched off Dnmt1. Methylation came to a halt at this point and after a short period of time, the genome was almost completely demethylated.
“Interestingly, when only Dnmt1 was switched back on, methylation levels began to rise despite there being no previous methyl marks to maintain”, Haggerty says. This was conclusive confirmation that the Dnmt1 enzyme can methylate DNA anew, or “de novo”.
Sequence analysis showed that Dnmt1 shows a particular preference for DNA regions that contain a certain type of transposon, which is common among rodents. The “Intracisternal A Particles” (IAP) are retrotransposons whose life cycle is strikingly similar to retroviruses such as the human immunodeficiency virus (HIV).
The cell reads the retroviral sequence from its DNA and produces RNA molecules that contain the blueprint for proteins. Among these is an enzyme that turns RNA back into DNA and another that inserts this DNA back into the cell's genome.
In fact, Dnmt1 very specifically targets IAP sequences in the genome of the mouse cells, Haggerty says: “It methylated some sections up to 18 percent – a very high level for an enzyme that was not expected to have that ability.”
The researchers have already discovered a few clues about the way Dnmt1 seeks out its target. In cell culture-based experiments, they identified other proteins that seem to attach to the Dnmt1 enzyme and that are known to interact with retrotransposons.
In studying the retrotransposons, the scientists quickly reached the limits of conventional technology. “We examine relatively short DNA fragments in traditional sequencing,” says computational scientist Helene Kretzmer, also a first author on the paper. “We look for overlaps and similarities in the sequences and piece them together. Because the retrotransposon copies that are scattered throughout the genome share a similar sequence, it's often hard to tell exactly which region we are looking at.”
Consequently, the researchers used a sequencing method in which the DNA strand is cut into relatively long segments that are then threaded through nanometer-sized pores on a chip. “Conventional methods give us fragments of about 150 to 300 bases, but nanopore sequencing spans the entire transposon with derived fragments of 10,000 to 30,000 bases,” Kretzmer says. “For methylation analysis of repetitive sequences, this method is currently unparalleled.”
“Of course, the main task of Dnmt1 is undoubtedly still to maintain methylation,” Meissner says. “Looking across the genome, the global de novo activity of the enzyme is low, but we showed that in specific contexts it can be more significant.”
Recently, other research groups had found increasing evidence that Dnmt1 is able to methylate DNA on its own to a small extent. The current study substantiates this suspicion. “We have put an end to years of debate and show that some classifications of the DNA methyltransferases may need to be revisited,” Meissner says. “At the same time, we are opening a new chapter by asking: in what other contexts could Dnmt1’s de novo activity have biological relevance?"
Original publication
Haggerty C, Kretzmer H, Riemenschneider C, Kumar AS, Mattei AL, Bailly N, Gottfreund J, Giesselmann P, Weigert R, Brändl B, Giehr P, Buschow R, Galonska C, von Meyenn F, Pappalardi MB, McCabe MT, Wittler L, Giesecke-Thiel C, Mielke T, Meierhofer D, Timmermann B, Müller FJ, Walter J, Meissner A; "Dnmt1 has de novo activity targeted to transposable elements"; Nat Struct Mol Biol.; 2021 Jun 17.