Tracking RNA through space and time
Completely new avenues for studying such things as the mechanism of action of drug therapies
A research team at the MDC has succeeded in tracking genes through space and time within a one-cell zebrafish embryo – even before cell division occurs. They have now described a method in the journal “Nature Communications” that may one day allow scientists to measure cell response to drugs, for example, in organoids.
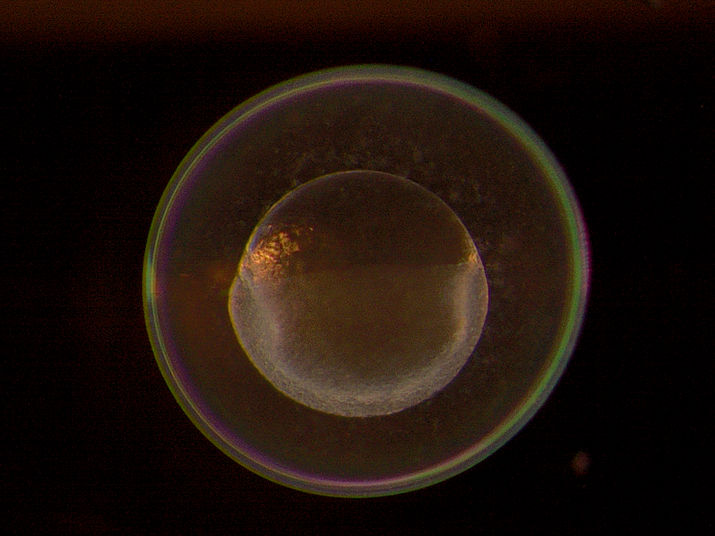
One-cell zebrafish embryo: The MDC research lab found numerous localized genes at this early stage of development. Much of their genetic information flows into the precursor cells of the later germ cells
© AG Junker, MDC
The “miracle of life” is most obvious at the very beginning: When the fertilized egg cell divides by means of furrows into blastomeres, envelops itself in an amniotic sac, and unfolds to form germ layers. When the blastomeres begin to differentiate into different cells – and when they eventually develop into a complete organism.
“We wanted to find out whether the later differences between the various cells are already partly hard-wired into the fertilized egg cell,” says Dr. Jan Philipp Junker, who heads the Quantitative Developmental Biology Lab at the Berlin Institute for Systems Biology (BIMSB) of the Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC). Junker and his team are investigating how cells make decisions and what dictates whether they become nerve, muscle, or skin cells. This involves creating cell lineage trees that allow them to determine the lineage and cell type of thousands of individual cells from an organism. Using these lineage trees, they can understand how and by what mechanisms cells come together to form a functioning organism or how they respond to perturbations.
Blueprints for different cell types already exist in the one-cell embryo
Yet this search for clues by means of cell lineage trees begins at a later stage – namely, when cell division and differentiation is already under way. What’s more, the observations cover long time periods. In their current study Junker and his team focus on a very short time period: the first hours after fertilization, from the one-cell stage to the process of gastrulation – the formation of the germ layers – of the embryo. The scientists wanted to know whether the one-cell embryo already contains parts of the blueprint for the multitude of different cell types that later develop from it. To do this, they studied zebrafish and clawed frog embryos. Researchers had previously succeeded in finding individual genes whose RNA is localized at specific sites within one-cell zebrafish embryos. The Berlin scientists have now shown that there are many more such genes. “We have discovered ten times more genes whose RNA is spatially localized in the fertilized egg cell than previously known,” explains Karoline Holler, lead author of the study. “Many of these RNA molecules are later transported into the primordial germ cells. This means that the program for subsequent cell differentiation is hard-wired into the fertilized egg cell.”
New approaches in transcriptomics
State-of-the-art methods of single-cell transcriptomics provide a good understanding of cell differentiation. Scientists order individual cells according to the similarity of their transcriptome – the complete collection of RNA molecules present in a cell – and can use the patterns that emerge to decipher how the cells became what they are. However, they cannot use this method to reconstruct the earliest stages of embryonic development, because here the spatial arrangement of RNA
molecules is crucial. His team instead used a specialized technique called tomo-seq, which Junker developed at the Hubrecht Institute in the Netherlands in 2014. It enables scientists to spatially track RNA molecules within the cell. This is achieved by cutting embryos of the model organisms into thin slices. It is then possible read the RNA profiles on the cut surfaces and convert them into spatial expression patterns. Holler refined the tomo-seq technique to now measure the spatial distribution of the transcriptome within the fertilized egg cell.
The scientists used another new technique to study which localized genes later contribute to which cells. “We labeled the RNA molecules so as to be able to track them over different developmental stages. This allows us to observe the RNA not only in space but also over time,” explains Junker. In this way, the scientists can distinguish the RNA transferred to the embryo by the mother from the RNA produced by the embryo itself. This RNA labeling method, called scSLAM-seq, was fine-tuned at BIMSB in the labs of Professor Markus Landthaler and Professor Nikolaus Rajewsky, enabling it to be applied in living zebrafish. “Labeling RNA molecules allows us to measure with high precision how gene expression changes in individual cells, for example, after an experimental intervention,” explains Junker.
How do drugs affect cell differentiation?
RNA labeling opens up completely new avenues for studying such things as the mechanism of action of drug therapies. “We can use it in organoids to investigate how different cell types respond to substances,” explains the physicist. The method, Junker says, is not suitable for long-term processes of change. “But we can see which genes change within five to six hours after treatment, providing a pathway to understanding how we might influence cell differentiation.”
Spatial analysis also has medical relevance: Looking further into the future, it could be useful for studying those diseases that result from mislocalized RNA, such as cancer or neurodegenerative diseases. In such diseases a large number of molecules are transported through the cell. “If we understand these transport processes, then we may be able to identify risk factors for these diseases,” explains Holler. But, for now, that is a long way off. “There is still much work to be done before the one-cell zebrafish embryo can be used as a model system for studying human neurodegenerative diseases,” stresses Junker.
The scientists next want to uncover the mechanisms involved in RNA localization: How does the detected RNA differ from other transcripts in the cell? Junker’s team plans to work with Professor Irmtraud Meyer’s lab at BIMSB to characterize the sequence features of the localized RNA. With the help of algorithms, they hope to predict whether the localized genes share a two- or three-dimensional fold. They are also working on further developing their method so that it can be used in other systems than the one-cell zebrafish embryo.