Watching cell division live
Highest resolution insights into living cells
Bacteria as unicellular organisms normally reproduce by binary cell division, i.e. the duplication of the entire organism consisting of a single cell. This allows particularly rapid multiplication, such as the exponential growth known from pathogens, and requires the separation and distribution of two identical copies of the bacterial genetic material. An important factor in cell division for a bacterial cell in this process is the position of its division, distributing the genetic information. For many bacterial species, such as the rod-shaped bacterium Escherichia coli, this process has been well researched: here, a group of proteins known as the Min system acts as a control unit and ensures that the cells divide exactly in the middle. At Kiel University, the Microbial Biochemistry and Cell Biology group led by Professor Marc Bramkamp is working on bacterial organization and reproduction mechanisms, among other things, which they are using to investigate generally applicable principles of biological pattern formation. The latter is of central importance in biology, for example in the development of complex multicellular organisms.
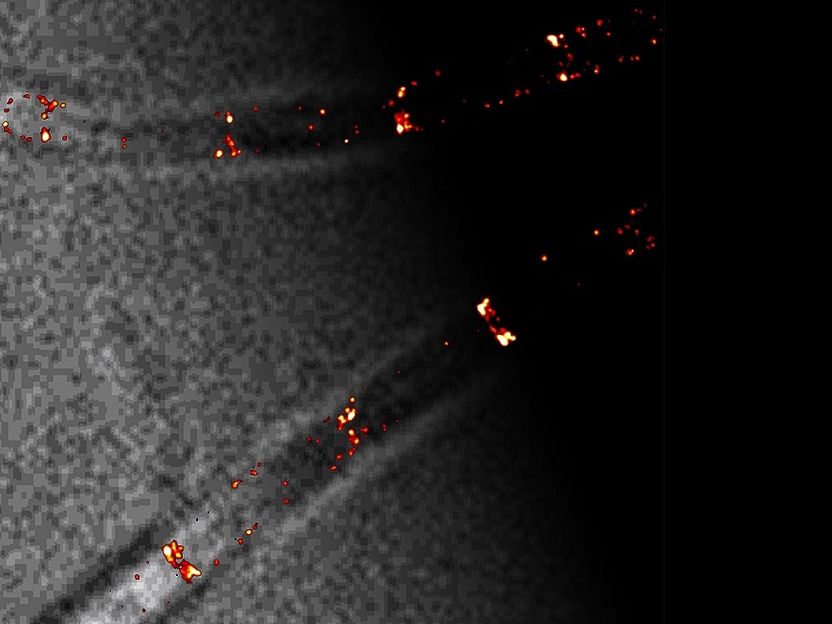
Certain fluorescently labeled proteins of the Min system (in red) can be seen at their positions in a B. subtilis cell. The protein localizations achieved by single-molecule localization microscopy are about ten times more accurate than conventional light microscopy.
© Prof. Marc Bramkamp
In a new research paper, Bramkamp and his team from the Institute of General Microbiology at Kiel University used the bacterium Bacillus subtilis as an example to gain new insights into how, as a result of evolutionary adaptations, the proteins of the Min system can adjust their function during cell division. Thanks to novel high-resolution imaging techniques and the application of mathematical modelling, they were able to show that the Min system in B. subtilis does not regulate the positioning of cell division, but stops it after successful division - unlike in many other rod-shaped bacteria. The new results were published by the Kiel researchers together with collaborators at Ludwig-Maximilians-University (LMU) in Munich in the scientific journal mBio.
Different dynamics of cell division proteins
In order to understand how the proteins of the Min system act in B. subtilis cells, the Kiel research team initially chose a two-step approach. Results from previous experiments suggested that the proteins involved in cell division are not static, but move dynamically in the cell depending on their state of activity. The researchers therefore first marked them with a fluorescent protein in the experiment. Using a specially equipped fluorescence microscope, they were then able to follow the proteins' movements in the cell in real time. "In E. coli, there is a so-called oscillation of Min proteins. In this process, their presence moves from one end of the cell to the other in a few seconds. Therefore, the average distribution is lowest in the exact centre of the cell. This is exactly where the septum, a newly forming partition wall, is formed in E. coli," explains Dr Helge Feddersen, a research associate in Bramkamp's group. "In B. subtilis, however, we could not detect such an oscillation with our measurements. However, our observations showed that Min proteins move from the cell poles to the centre right after cell division begins there. So, while protein dynamics are preserved in B. subtilis, the system does not determine the site of division," Feddersen added.
In order to get a precise idea of the speed of the proteins moving in the cell, the Kiel team applied a method for local removal of the fluorescent label in a second step. The fluorescence was thus permanently switched off, for example for the proteins located at the division septum of the cell, by bleaching with a laser. Nevertheless, fluorescent proteins were found again shortly thereafter at the same position - these could therefore only have migrated there from another location in the cell. "From this, we were able to deduce quite precisely the speed at which the Min proteins move in the cell in B. subtilis. These quantitative data are very important to build a mathematical model of Min dynamics," Feddersen summarizes the most important result. To confirm the processes observed in the living cells, the Kiel researchers collaborated with the research group of physicist Professor Erwin Frey at LMU. In a theoretical model, the Munich cooperation partners were also able to theoretically confirm the movement patterns of the Min system - an important sign that all parts of the protein system essential for its function were already identified in the observations.
Highest resolution insights into living cells
The fact that the new insights into the deviant function of the Min system in B. subtilis have only now been found is also due to the fact that the resolution in imaging was previously insufficient for such observations. At Kiel University, single-molecule localization microscopy has recently become available, a high-performance technology that provides the best resolution yet in living cells. This makes it possible to localize single molecules such as Min proteins in living cells on the nanometre scale. In contrast to static methods, this also makes it possible to track their dynamics, i.e., the movement of individual proteins over time. "It was only with our highest-resolution microscope that we were now able to determine in the next step that clustering occurs in the cell division proteins of B. subtilis," emphasizes Bramkamp, head of the Microbial Biochemistry and Cell Biology group. "With the new microscopy technology, Kiel University is now excellently positioned in biological imaging. Within the framework of the Central Microscopy facility at the Biology Centre, it is also available to other users from diverse disciplines at our university," says Bramkamp who is also scientific head of the imaging facility.
"This technique, which is still rarely used in biology, has shown us that an apparently well-known protein system may function much differently in the specific bacterium studied than previously assumed," Bramkamp continues. So, although the same protein systems are present in E. coli and B. subtilis, for example, they perform completely different functions. "We suspect that the Min system in B. subtilis causes cell division to be blocked after a successful passage - in other words, it influences the active cell division apparatus and ensures that it does not continue to work after successful division," Bramkamp summarizes. The reason for this adaptation may be that B. subtilis pursues a special survival strategy and - in contrast to E. coli, for example - forms permanent states in the form of spores that are resistant to harmful environmental influences. In order for these spores to form, the cell must divide close to the cell pole - i.e. the site where division should normally not take place. Thus, B. subtilis apparently achieves this requirement by modifying the Min system, thus enabling this simple form of cell differentiation during sporulation.