New method reveals how the Parkinson's disease protein damages cell membranes
The method makes it possible to study tiny quantities of biological molecules without using fluorescent markers
In sufferers of Parkinson's disease, clumps of α-synuclein (alpha-synuclein), sometimes known as the 'Parkinson's protein', are found in the brain. These destroy cell membranes, eventually resulting in cell death. Now, a new method developed at Chalmers University of Technology, Sweden, reveals how the composition of cell membranes seems to be a decisive factor for how small quantities of α-synuclein cause damage.
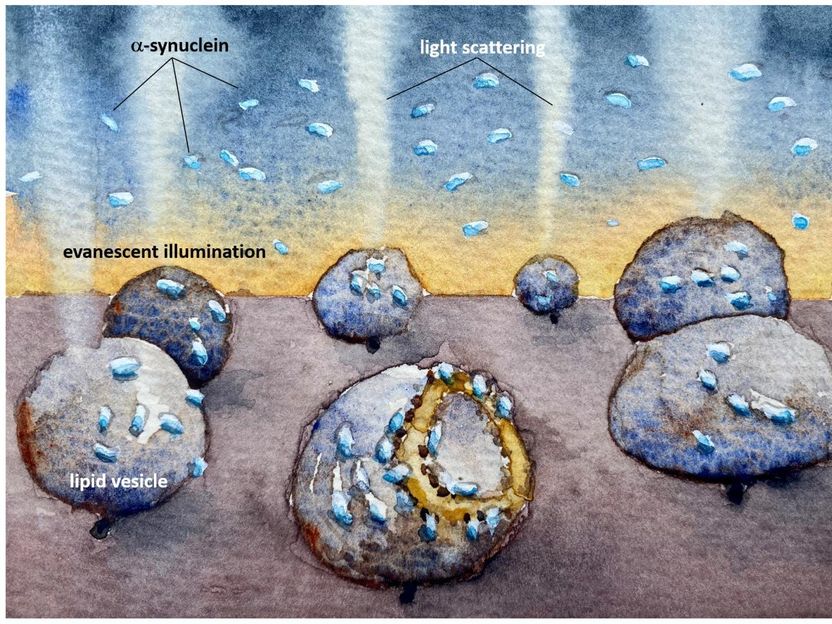
An illustration showing how the mitochondria-imitating lipid vesicles are damaged by the Parkinson's protein alpha synuclein. The light scattering reveals how the membrane is destroyed even at very low, nanomolar, concentrations, where proteins do not aggregate to clumps before they bind.
Fredrik Höök/Chalmers University of Technology
Parkinson's disease is an incurable condition in which neurons, the brain's nerve cells, gradually break down and brain functions become disrupted. Symptoms can include involuntary shaking of the body, and the disease can cause great suffering. To develop drugs to slow down or stop the disease, researchers try to understand the molecular mechanisms behind how α-synuclein contributes to the degeneration of neurons.
It is known that mitochondria, the energy-producing compartments in cells, are damaged in Parkinson's disease, possibly due to 'amyloids' of α-synuclein. Amyloids are clumps of proteins arranged into long fibres with a well-ordered core structure, and their formation underlies many neurodegenerative disorders. Amyloids or even smaller clumps of α-synuclein may bind to and destroy mitochondrial membranes, but the precise mechanisms are still unknown.
The new study, recently published in the journal PNAS, focuses on two different types of membrane-like vesicles, which are 'capsules' of lipids that can be used as mimics of the membranes found in cells. One of the vesicles is made of lipids that are often found in synaptic vesicles, the other contains lipids related to mitochondrial membranes.
The researchers found that the Parkinson's protein would bind to both vesicle types, but only caused structural changes to the mitochondrial-like vesicles, which deformed asymmetrically and leaked their contents.
"Now we have developed a method which is sensitive enough to observe how α-synuclein interacts with individual model vesicles. In our study, we observed that α-synuclein binds to - and destroys - mitochondrial-like membranes, but there was no destruction of the membranes of synaptic-like vesicles. The damage occurs at very low, nanomolar concentration, where the protein is only present as monomers - non-aggregated proteins. Such low protein concentration has been hard to study before but the reactions we have detected now could be a crucial step in the course of the disease," says Pernilla Wittung-Stafshede, Professor of Chemical Biology at the Department of Biology and Biological Engineering..
The new method from the researchers at Chalmers University of Technology makes it possible to study tiny quantities of biological molecules without using fluorescent markers. This is a great advantage when tracking natural reactions, since the markers often affect the reactions you want to observe, especially when working with small proteins such as α-synuclein.
"The chemical differences between the two lipids used are very small, but still we observed dramatic differences in how α-synuclein affected the different vesicles," says Pernilla Wittung-Stafshede.
"We believe that lipid chemistry is not the only determining factor, but also that there are macroscopic differences between the two membranes - such as the dynamics and interactions between the lipids. No one has really looked closely at what happens to the membrane itself when α-synuclein binds to it, and never at these low concentrations."
The next step for the researchers is to investigate variants of the α-synuclein protein with mutations associated with Parkinson's disease, and to investigate lipid vesicles which are more similar to cellular membranes.
"We also want to perform quantitative analyses to understand, at a mechanistic level, how individual proteins gathering on the surface of the membrane can cause damage" says Fredrik Höök, Professor at the Department of Physics, who was also involved in the research.
"Our vision is to further refine the method so that we can study not only individual, small - 100 nanometres - lipid vesicles, but also track each protein one by one, even though they are only 1-2 nanometres in size. That would help us reveal how small variations in properties of lipid membranes contribute to such a different response to protein binding as we now observed."