Bacteria leave signature in colon cancer cells
Scientists identify mutations in the genome caused by the intestinal bacterium Escherichia coli
Advertisement
Some bacterial pathogens cause damage in the genomes of their infected cells which could lead to the initiation of cancer. While it is difficult to link an infection with an onset of cancer that arises many years later in life, researchers have been looking for definitive proof that such links really exist – apart from the available epidemiological evidence and mechanistic plausibility. One strong argument for the causative role of an infection in cancer would be the identification of a footprint, a genetic signature that bacteria leave in the genomes of their infected cells and can be detected in the transformed cancer cells. This is what researchers under the lead of Thomas F. Meyer, Director at the Max Planck Institute for Infection Biology in Berlin, together with colleagues from Helsinki, Stockholm, and Barcelona now identified. They observed a specific signature of E. coli bacteria in a subgroup of colon cancer patients, and thereby provided first definitive evidence for an etiologic link between a bacterium and cancer development.
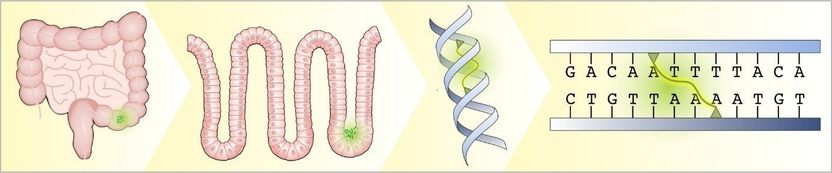
Escherichia coli bacteria nest in crypts of the intestinal epithelium. They release a toxin into the epithelial cells, which binds to the DNA strands of the cells and links them together.
© MPI f. Infection Biology
Starting from the hypothesis that DNA damage by the colibactin genotoxin in the human genome might not be random, the authors determined the locations of double strand breaks generated after the infection of human cells with colibactin-positive E. coli. They found a strong enrichment of breaks at a distinct sequence motif that exhibited extreme structural features. Computer modelling indicated that the target motif was in a particularly narrow groove area in the DNA, to where usually no proteins bind. This motif was rich of adenine:thymidine (A:T) base pairs and it nicely fitted the binding by colibactin. Accordingly, colibactin reacted with two adenines on opposite strands of the DNA, cross-linking the two DNA strands at a diagonal distance of 3 nucleotides.
Enrichment of CDM-associated mutations
When the researchers explored whether this colibactin damage motif (CDM) was mutated in human cancers, they found a strong enrichment of CDM-associated mutations in a subgroup of human colorectal cancers. Moreover, the position where colibactin caused the damage in the target DNA, i.e. where it reacted with the two adenines, corresponded exactly with the location of the mutations in the colon cancer genomes. Obviously, in the course of repairing the affected DNA, the human cell happens to make occasional mistakes by incorporating a mismatched nucleotide. Interestingly, the researchers noted that this faulty rescue has a slight bias towards the incorporation of a guanine or a cytosine in the respective DNA strands.
These observations point to a highly specific mutational signature, termed SBSA, which has been recently noted in another study to be present in seemingly healthy intestinal tissue. ‘Both our own and published data indicate that these colibactin-specific mutations happen early in life’ says Hilmar Berger, the bioinformatician in the Max Planck team. ‘ who did most of the bioinformatics analyses, obviously being intrigued by this unexpected conclusion. ‘Perhaps there exists a particularly vulnerable phase in childhood or early youth’ Paulina Dziubanska-Kusibab adds.
Mutations in tumor suppressor gene
The researchers also asked whether the mutations caused by colibactin have an impact on carcinogenesis. In colon cancer, mutations in the tumor suppressor APC usually occur first as they tend to confer independence of certain growth factors. In fact, the Max Planck team noted that, amongst the mutated genes showing a colibactin signature, the APC gene was frequently affected. ‘This strongly corroborates the causative role of colibactin in colon cancer’, Meyer says and continues: ‘The new data urge for a rethinking on the use of colibactin-producing E. coli bacteria as probiotics. Still, we need to learn more about the physiological conditions, under which colibactin-producing bacteria could cause mutations and whether only certain people and age groups are affected.’
Interestingly, other bacteria were also shown to carry the genomic island responsible for colibactin synthesis, or produce other genotoxins that cause cancer-promoting mutagenesis. This opens new avenues of research into the interaction of the microbial genotoxins and epithelial cells and their role in carcinogenesis.
Original publication
Paulina J. Dziubańska-Kusibab, Hilmar Berger, Federica Battistini, Britta A. M. Bouwman, Amina Iftekhar, Riku Katainen, Nicola Crosetto, Modesto Orozco, Lauri A. Aaltonen, Thomas F. Meyer; "Colibactin DNA-damage signature indicates mutational impact in colorectal cancer"; Nature Medicine; June 01, 2020