Looking for new antibiotics
DSMZ scientist develops innovative strategy for drug screening
Advertisement
A team of researchers led by Professor Dr. Yvonne Mast from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures in Braunschweig (Germany) have developed a new strategy for the activation of silent antibiotic gene clusters. They showed that the Streptomyces Antibiotic Regulatory Protein (SARP), a regulator that controls the biosynthesis of antibiotics in streptomycetes, can be used as a general activator of various antibiotic gene clusters. The research data on how SARPs can be used to identify and specifically switch on inducible gene clusters have been published in the journal Frontiers in Microbiology.
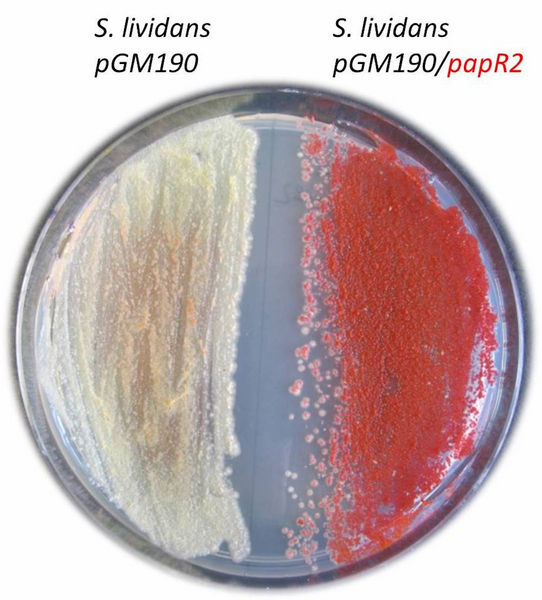
Growth of Streptomyces lividans on agar plate. On the left the control, on the right the strain with the SARP overexpression construct (pGM190/papR2).
DSMZ/Mast
In their study, the team of Prof. Yvonne Mast was able to show that SARP regulators occur in many different actinomycetes and are part of gene clusters that code for different types of secondary metabolites. In their experiments the researchers showed that SARP regulators have the potential to regulate gene clusters also in other host strains. Using the SARP regulator PapR2 as an example, the researchers demonstrated that this regulator has cluster-activating properties not only in the bacterium Streptomyces pristinaespiralis, in which it occurs naturally; it is also able to activate silent gene clusters in other streptomycetes such as Streptomyces lividans, thereby enabling the production of bioactive substances. By better understanding regulatory mechanisms and with the help of genome sequence data, researchers can now predict which gene clusters can potentially be activated by SARPs, which will facilitate later drug discovery.
Activation of silent gene clusters
Actinomycetes are known for their potential to produce many bioactive substances. Based on genome sequence analyses, it is known that the organisms harbour numerous so-called "silent gene clusters", which contain the information for the synthesis of active substances, which however is not translated into the corresponding substances. It is assumed that actinomycetes theoretically could produce ten times more secondary metabolites than are normally isolated under standard laboratory conditions. Why many of the clusters are "inactive" is still unclear. However, one reason might be the lack of activation by regulatory elements. The discovery and activation of such silent gene clusters, which may result in the identification and characterisation of new potential drugs are of utmost importance in times of increasing numbers of infections caused by antibiotic-resistant pathogens.
"The advantage of the method we use is that it allows us to activate gene clusters in a relatively simple but targeted manner. Most other strategies for the activation of silent gene clusters are either unspecific activation approaches, which require major analytical efforts or are gene cluster-specific, which involves a lot of cloning work." Professor Yvonne Mast summarizes the advantages of her research approach.