Antibiotic intercepts building blocks of the bacterial envelope
One of the last arrows in the quiver in the fight against dangerous bacteria is the reserve antibiotic daptomycin. It is used primarily when conventional drugs fail due to resistant bacteria. Although the antibiotic was developed around 30 years ago, its exact mode of action was previously unclear. Scientists at the University of Bonn have now deciphered the puzzle: Daptomycin blocks the integration of important building blocks into the cell wall of the pathogens, thereby killing the bacteria.
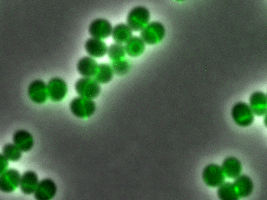
Daptomycin binds to special areas of the cell envelope of the bacterium Staphylococcus aureus, which are rich in attacking structures. The fluorescence-labelled daptomycin glows green. These are the areas where the cells are currently dividing.
© Fabian Grein
When it comes to bacterial infections, antibiotics are the medical weapons of choice - but they are becoming increasingly blunt. The number of resistances is increasing, which is why many antibiotics are no longer effective against the dangerous pathogens. Some of these active substances are intended for particularly severe infections with resistant bacteria. One example is daptomycin, which was launched in the USA in 2003 and in Germany in 2006. It is used for the treatment of infections with methicillin-resistant Staphylococcus aureus (MRSA) and resistant enterococci.
"Although daptomycin was discovered around 30 years ago, the exact mode of action remained elusive until now," explains Prof. Dr. Tanja Schneider from the Institute of Pharmaceutical Microbiology at the University of Bonn and the German Center for Infection Research (DZIF). There were different theories on how this antibiotic attacks and kills bacteria. One of them was that daptomycin perforates the bacterial envelope and leads to a potassium efflux that ends with the death of the bacterium. "Nobody understood how daptomycin actually works," says Dr. Anna Müller, one of the lead authors from Prof. Schneider's research group.
Interdisciplinary research team
The interdisciplinary team from the fields of medicine, pharmacology and physical chemistry used a wide variety of scientific methods to discover the antibiotic's mode of action. The researchers first labelled daptomycin with a fluorescent dye that glows green. This enabled them to follow exactly where the antibiotic docks to the staphylococcal cells under the high-resolution microscope. "Daptomycin binds to the bacteria in regions where the new cell wall is just being synthesized," said Dr. Fabian Grein, another lead author and colleague of Prof. Schneider.
Like in a construction kit, the bacterial cell wall is assembled from numerous building blocks. Further analyses carried out by the researchers on staphylococci and synthetically produced bacterial walls showed that two of these building blocks in particular are hugely important for the effect of daptomycin: the central cell wall building block "lipid II" and the membrane lipid phosphatidylglycerol (PG). "The combination of lipid II and PG together is the Achilles' heel of the bacteria," says Schneider. This is exactly where daptomycin comes into play: The antibiotic captures these important building blocks and blocks the further construction of the cell wall. As a result, the bacterial cell wall becomes unstable resulting in the outflow of various ions, including potassium. "The outflow of ions is not the actual killing mechanism of daptomycin, as originally thought, but a consequence of bacterial cell death," concludes Schneider.
"We were able to show how daptomycin really works and to which molecular target structures it docks," comments Prof. Dr. Ulrich Kubitscheck from the Department of Biophysical Chemistry at the University of Bonn. This is an important prerequisite for the further optimization of daptomycin. Since new active substances cannot be developed to the desired extent to fight antibiotic resistance, researchers are focusing on combination therapies using different active substances. "The strategy is to target already resistant bacteria with differently acting weapons," says Prof. Schneider. However, this would only work if the mode of action and targets of the antibiotics are known.
The study was conducted in the Transregional Collaborative Research Center TRR261 "Antibiotic CellMap - Cellular Mechanisms of Antibiotic Action and Production" located at the Universities of Bonn and Tübingen and funded by the German Research Foundation (DFG). The German Center for Infection Research is also involved. "It was only through this transdisciplinary cooperation that we were able to take the decisive step forward and solve a puzzle that science has been mulling over for 30 years," says Schneider.
Original publication
Fabian Grein, Anna Müller, Katharina M. Scherer, Xinliang Liu, Kevin C. Ludwig, Anna Klöckner, Manuel Strach, Hans-Georg Sahl, Ulrich Kubitscheck & Tanja Schneider; "Ca2+-Daptomycin targets cell wall biosynthesis by forming a tripartite complex with undecaprenyl-coupled intermediates and membrane lipids"; Nature Communication; 2020