Bristol pioneers use of VR for designing new drugs
Researchers at the University of Bristol are pioneering the use of virtual reality (VR) as a tool to design the next generation of drug treatments.

User interacting with a protein in VR.
University of Bristol
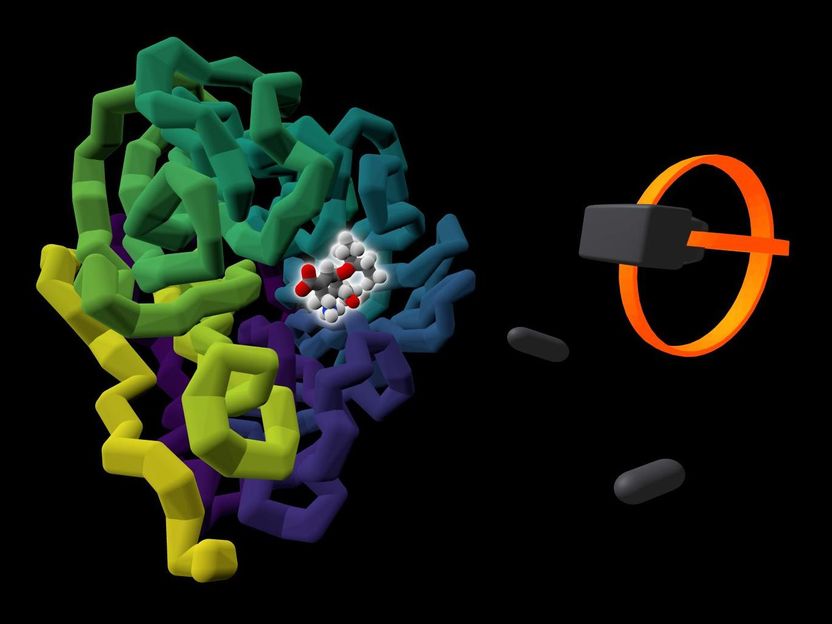
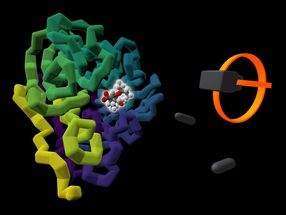
The commercially available influenza drug Tamiflu (highlighted by a glow) is shown bound to the viral protein it targets, neuraminidase (coloured in purple to yellow). A rendering of a user's VR headset and controllers is presented alongside the protein.
University of Bristol

The findings, published in the journal PLOS ONE describe how researchers used VR to understand how common medications work on a molecular level.
Many drugs are small molecules, and discovering new drugs involves finding molecules that bind to biological targets like proteins.
In the study, users were able to use VR to 'step inside' proteins and manipulate them, and the drugs binding to them, in atomic detail, using interactive molecular dynamics simulations in VR (iMD-VR).
Using this iMD-VR approach, researchers 'docked' drug molecules into proteins and were able to predict accurately how the drugs bind. Among the systems studied were drugs for flu and HIV.
Professor Adrian Mulholland, from the University of Bristol's Centre for Computational Chemistry, and co-lead of the work, said: "Many drugs work by binding to proteins and stopping them working. For example, by binding to a particular virus protein, a drug can stop the virus from reproducing.
"To bind well, a small molecule drug needs to fit snugly in the protein. An important part of drug discovery is finding small molecules that bind tightly to specific proteins, and understanding what makes them bind tightly, which helps to design better drugs.
"To design new therapies, researchers need to understand how drug molecules fit into their biological targets. To do this, we use VR to represent them as fully three-dimensional objects. Users can then fit a drug within the 'keyhole' of a protein binding site to discover how they fit together."
In the study, users were set the task of binding drugs to protein targets such as influenza neuraminidase and HIV protease.
Tests showed that users were able to predict correctly how the drugs bind to their protein targets. By pulling the drug into the protein, they could build structures that are very similar to the structures of the drug complexes found from experiments.
Even non-experts were able to dock drugs into the proteins effectively. This shows that interactive VR can be used to predict accurately how new potential drugs bind to their targets.
The study shows how VR can be used effectively in structure-based drug design, even by non-experts. It uses readily available VR equipment and an open source software framework, so can be applied by anyone.
Professor Mulholland added: "An important aspect of the work is that the drugs, and their protein targets, are fully flexible: we model their structural changes and dynamics, and users can manipulate them interactively to find how drugs interact with their biological targets. This is a really exciting and powerful way to model drug binding. We have shown in this work that it gives accurate results. These tools will be useful in the design and development of new drugs."
Dr David Glowacki, Royal Society Senior Research Fellow in Bristol's School of Chemistry and Department of Computer Science, said: "Our results show that it is possible to unbind and rebind drugs from protein targets on a simulation timescale significantly shorter than the timescale of similar events observed using non-interactive molecular dynamics engines.
"It is also important to note that the full unbinding and rebinding events generated using iMD-VR were achieved by the users in less than five minutes of real time.
"Where non-expert users had trace atoms showing them the correct pose, all participants were able to establish a docking pose which was close enough to the starting structure to be scientifically considered redocked.
"Where no trace atoms were present, binding poses understandably had more variation, but users were still able to get within the same range of the accepted bound position for all three systems. These results were achieved within a single hour-long training session with each participant, demonstrating the usability of this VR framework."
Original publication
H. Deeds, R. Walters, S. Hare, M. O’Connor, A. Mulholland and D. Glowacki; "Interactive molecular dynamics in virtual reality for accurate flexible protein-ligand docking"; PLOS One