Inner “clockwork” sets the time for cell division in bacteria
Advertisement
Researchers at the Biozentrum of the University of Basel have discovered a “clockwork” mechanism that controls cell division in bacteria. In two publications, in “Nature Communications” und “PNAS”, they report how a small signaling molecule starts the “clock”, which informs the cell about the right time to reproduce.
The signaling molecule c-di-GMP controls cell division in Caulobacter crescentus.
University of Basel, Swiss Nanoscience Institute/Biozentrum
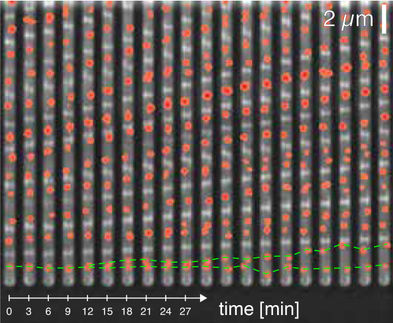
The ability of pathogens to multiply in the host is crucial for the spread of infections. The speed of bacterial division greatly depends on the environmental conditions. Under unfavorable conditions, such as nutrient deficiency, bacteria tend to pause after division and reproduce more slowly. But how do bacteria know, when it is time to enter the next round of cell division?
A team at the Biozentrum of the University of Basel, led by Prof. Urs Jenal has now identified a central switch for reproduction in the model bacterium Caulobacter crescentus: the signaling molecule c-di-GMP. In their current study, published in the journal Nature Communications, they report that this molecule initiates a “clock-like” mechanism, which determines whether individual bacteria reproduce.
A signaling molecule regulates “clockwork” in bacteria
How long a cell pauses after division and how it then decides to engage in the next round of division is still poorly understood. The signaling molecule c-di-GMP plays a key role in this process. “The rise in the c-di-GMP level sets the individual cogwheels of the cell’s clock into action, one after the other,” explains Jenal. “These cogwheels are enzymes called kinases. They prepare the transition of the cell from the resting to the division phase.”
Enzymes respond to c-di-GMP levels
Under favorable living conditions, newborn bacteria begin to produce the signaling molecule – this starts the clock ticking. The initially low c-di-GMP level activates a first kinase. This activates the expression of over 100 genes, which drive the cell towards division and boost the production of c-di-GMP.
The resulting peak levels of c-di-GMP finally stimulate the last wheel of the machinery, also a kinase. “With this step, the cell ultimately decides to replicate its DNA and to trigger cell division,” explains Jenal. “Simultaneously the over 100 genes are switched off again, as these are only important for the transition phase but obstruct later stages of proliferation.”
Insights into c-di-GMP mediated enzyme activation
In a parallel study, recently been published in PNAS, a team led by Prof. Tilman Schirmer, also at the Biozentrum of the University of Basel, describes how c-di-GMP activates the first cogwheel of the newly discovered clock at the atomic level.
The researchers have revealed that the mobile domains of the kinase are initially locked in a fixed position. The binding of c-di-GMP liberates the domains, thereby activating the kinase for gene expression. “In our study, we have discovered a new mode of c-di-GMP mediated activation,” says Schirmer. “Once again, we are fascinated by the diverse ʻstrategiesʼ of this small molecule to regulate biochemical processes.”
Universal principle in bacterial reproduction
The c-di-GMP regulated timing of the bacterial cell cycle by this signaling molecule seems to be a universal mechanism. The researchers assume that this mechanism enables bacteria to precisely coordinate growth and development. The elucidation of this novel mechanism also contributes to a better understanding of the growth of bacterial pathogens.
Original publication
Andreas Kaczmarczyk, Antje M. Hempel, Christoph von Arx, Raphael Böhm, Badri N. Dubey, Jutta Nesper, Tilman Schirmer, Sebastian Hiller and Urs Jenal; "Precise timing of transcription by c-di-GMP coordinates cell cycle and morphogenesis in Caulobacter"; Nature Communications; 2020.
Badri N. Dubey, Elia Agustoni, Raphael Böhm, Andreas Kaczmarczyk, Francesca Mangia, Christoph von Arx, Urs Jenal, Sebastian Hiller, Iván Plaza-Menacho, and Tilman Schirmer; "Hybrid histidine kinase activation by cyclic di-GMP–mediated domain liberation"; PNAS; 2020.