New insights into the evolution of proteins
Advertisement
How do bacteria manage to adapt to synthetic environmental toxins and, for example, to even develop strategies for using a pesticide and chemical warfare agent as food within less than 70 years? The evolutionary adaptations underlying such processes have now been studied in detail by an international team of researchers. The scientists looked at an enzyme that they had isolated from bacteria that had been exposed to pesticides in the vicinity of factories producing these pesticides. Using a novel combination of methods, the researchers found out which mutations had occurred in the gene over the years – and how these mutations led to biochemical changes that now enable the enzyme to cleave the pesticide.
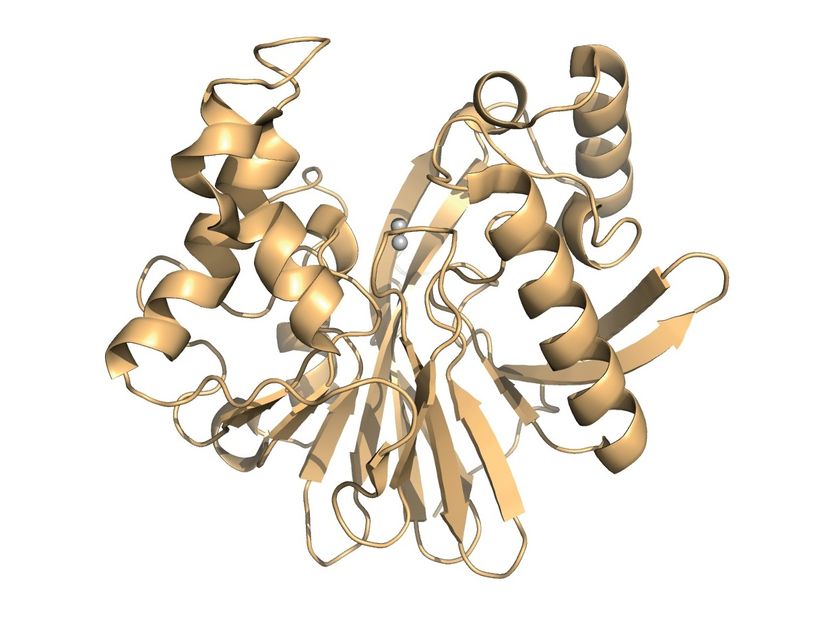
Model of the enzyme that the researchers investigated in their study. The two grey spheres represent the active centre that binds to the pesticide to cleave it.
G. Yang et al/Nat Chem Biol
"With our study, we show that the technique of the so-called ancestral reconstruction can not only be used to decipher evolutionary processes that date back many centuries, but also to investigate recent adaptations with very rapid evolution, such as those occurring in bacteria," emphasizes co-author Prof. Erich Bornberg-Bauer of the University of Münster, whose group was significantly involved in the study. The study was carried out in cooperation with the research group led by Prof. Nobuhiko Tokuriki from British Columbia University in Vancouver, Canada. Among other things, the results could help to find new ways to break down and dispose of environmental toxins and chemical substances.
Background and method
The researchers used the method of ancestral reconstruction, which uses algorithms to recover the state of proteins in ancestors. To do this, the scientists "resurrect" a kind of primordial gene that is regarded as the starting point of today's gene family. Classically, this method is used to reproduce proteins that change slowly over millions to billions of years. In this case, the researchers used this method to trace back a very rapidly evolving enzyme over a few decades and uncover molecular mechanisms that have led to a completely new function – the ability to cleave the pesticide methyl-parathion. It has only been produced industrially for about 70 years and is closely related to nerve toxins, which are also suitable for chemical warfare. The researchers combined the method of ancestral reconstruction with biochemical and structural tests of the “resurrected” protein in the laboratory and identified five mutations that are mainly responsible for the functional transition of the protein over time.
In a further step, the scientists investigated the extent to which these mutations changed the so-called fitness, i.e. the degree to which the genes adapted at certain points in time. In addition, they characterized many different combinations of the mutations. They used novel statistical analyses to determine the interaction of the genes and their different dominance. These analyses provided information on the underlying molecular interactions – both within the enzyme and between the enzyme and the pesticide.
The researchers found that the five most important of the identified mutations play together in a complex network and ultimately enable the active centre of the enzyme to connect better with the pesticide and thus develop new functions. "Our combined method can also be used in many other cases to find out more about the molecular mechanisms underlying the evolution of proteins," said co-author Elias Dohmen from the University of Münster, who reconstructed the ancestral protein sequences for the study as part of his PhD thesis in the Molecular Evolution and Bioinformatics group.
In future investigations, the scientists plan to test more proteins with different functions in order to derive generally usable rules from the methodology. In addition, they want to collect even more sequence data in order to be able to create better ancestral reconstructions.
The newly gained knowledge is relevant for the so-called directed evolution – a process that aims at the optimization of enzymes and proteins and is mainly used in industrial biotechnology. In 2018, Frances Arnold of the California Institute of Technology was awarded the Nobel Prize in Chemistry for this method.
In addition to the British Columbia University in Vancouver and the University of Münster, the Universities of Canberra in Australia and Uppsala in Sweden participated in the study.