DNA repair: Opening the hatch to heal the break
LMU researchers have determined the structure of a key enzyme complex that is involved in DNA repair, and traced the cycle of conformational changes that it undergoes while performing its biochemical function.
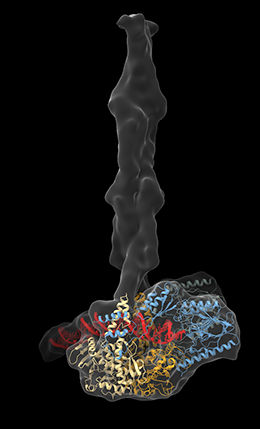
MR Complex.
L. Käshammer
Various types of DNA damage can have serious repercussions, both for the individual cells in which they occur and for the organism as a whole. Instances of simultaneous breakage of both strands of the DNA helix are particularly harmful. Such double-strand breaks (DSBs) can be induced by radiation, as well as by environmental toxins, and can lead to cell death. However, cells possess efficient mechanisms for detecting and repairing DSBs. A molecular machine known as the MR complex plays a central role in this process. It recognizes and binds to DSBs, and initiates repair of the broken double helix. A team of scientists led by Professor Karl-Peter Hopfner, who holds the Chair of Structural Molecular Biology at LMU’s Gene Center, has now deciphered the complete three-dimensional structure of the MR complex and determined how it works.
Hopfner’s group had previously shown that the MR complex is made up of four proteins. Two of these are nucleases, which themselves cut DNA strands. The others are ATPases – enzymes that remove a phosphate group from the molecule ATP, and in so doing release chemical energy. The complex itself has an open architecture. But when it encounters a DSB, it binds to the DNA and brings the ends together, acting as the molecular equivalent of a hook-and-loop fastener. The complex then detaches any chemical adducts that block the ends of the break and removes further subunits from the DNA. These operations then provide a ‘clean break’ in preparation for the repair process itself. “Because of the intricate architecture of the complex, the conformational changes involved in this process had not been elucidated,” says Lisa Käshammer, lead author of the paper. The ATPase enzymes contain prominent filamentous structural elements that are referred to as coiled-coils, which are made up of closely apposed and helically ordered amino-acid sequences. These filaments are long and flexible, and their mobility had made it impossible to discern their precise courses using conventional X-ray crystallographic methods.
The team turned to cryo-electron microscopy to get around this problem, and succeeded in determining the complete structure of the MR complex found in the bacterium Escherichia coli. The bacterial complex is somewhat less complicated than its counterpart in higher organisms, but in their overall structure the two forms are similar to each other. The new structure now makes it possible to discern how the nucleases actually bind to the DNA – a question that had remained open up to now. “In the previously published structures, the DNA was either positioned relatively far from the active center of the nuclease, or bound in such a way that the active center was inaccessible,” Käshammer explains. The new structure reveals that binding to the DNA causes the nuclease to execute a 120-degree turn, which opens up a channel that enables the DNA to bind to the active center of the enzyme.
The study also clarifies the function of the coiled-coil regions. As the new structural model demonstrates, these segments form a brace that stabilizes the DNA, and are actively involved in the binding and processing of the DNA ends. “Our findings constitute an important advance in the quest for a deeper understanding of the complex mechanisms that make it possible for these enzymes to repair double-strand breaks in DNA,” says Hopfner. New insights into the function of the complex are also of potential medical significance, as DSBs play a significant role in the pathogenesis of many types of cancer.
Original publication
Other news from the department science

Get the life science industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.
More news from our other portals
Last viewed contents
ATP as trigger for targeted anti-cancer drug delivery
Society for Neuroscience - Washington, USA
Pronova BioPharma's PRC-4016 to begin clinical trials for mixed dyslipidemia
Novozymes and Prometic enter into alliance for purification of albumin-fusion proteins
Salmonella Detection - Simple Colorimetric Sensor Rapidly Detects Food Contamination Using a Nucleic Acid Probe
Eisai Opens Regional Office in Bahrain
Pfizer Acquires Antibody Discovery and Optimization Company

Discovery of a potential new therapy for inflammatory arthritis - Researchers at the Schroeder Arthritis Institute in Toronto have made a discovery that could lead to new treatments for axial spondyloarthritis.

Lung organoids unveil secret: How pathogens infect human lung tissue - New sensor for monitoring bacteria

How genes and small molecules influence our personal disease risk - Metabolism also determines drug effects

Dangerous plastics - Ingestion of microplastics can trigger evolutionary changes