AI to Predict the Protein Structure
proteins are biological high-performance machines. They can be found in every cell and play an important role in human blood coagulation or as main constituents of hairs or muscles. The function of these molecular tools is obvious from their structure. Researchers of Karlsruhe Institute of Technology (KIT) have now developed a new method to predict this protein structure with the help of artificial intelligence.
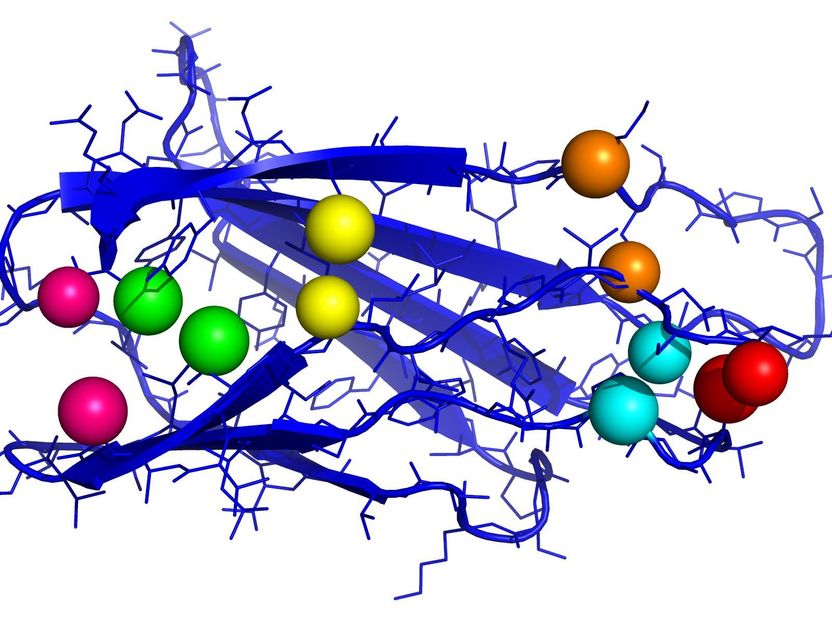
Fibronectin plays an important role in wound healing. The figure shows an important part of the protein with contact pairs (spheres of the same color).
Ines Reinartz, KIT
Depending on their structure, proteins can interact with other molecules by penetrating or enclosing them. This is very difficult to detect, the experiments needed for this purpose are expensive and complex. Researchers of the Steinbuch Centre for Computing (SCC), the computing center of KIT, have searched the databases for protein sequences and compared the same proteins of different species. “Hemoglobin that is responsible for transporting oxygen in our body can also be found in insects, voles, and chimpanzees,” says Markus Götz, data analyst of SCC. The protein structure resembles a string of pearls, with the string consisting of the protein components, the amino acids. Its three-dimensional structure and the associated properties result from some distant “pearls” forming pairs, thus folding the protein. These pairs may differ in different organisms. The properties of the protein, however, remain the same. “Harmful mutations are sorted out in the course of evolution,” Götz says.
Now, Götz’s research team has taught an artificial intelligence (AI) system which pairs proved to be successful in known protein sequences during evolution. “We expect the system to draw conclusions with respect to the structure of unknown protein sequences as well,” Götz says. The benefit: “It is easy to determine the amino acids which form the protein chain. However, it is very complex and costs millions to directly determine protein structures experimentally,” Alexander Schug, SCC, adds.
The use of AI to predict contacts in proteins is not new. “Currently, image processing methods are applied for this purpose,” Götz says. Such neural networks can recognize patterns well. When determining the protein structure, however, contacts of protein components located far away from each other are of crucial importance, because they have a stronger impact on the structure during folding than those that are located closely to each other. “For this reason, we use an approach from automatic language translation. We consider the amino acid chains sentences that have to be translated into another language.” So-called “self-attention neural networks” are applied in popular translation programs. They can identify which parts of the sentence are linked or, in the protein context, which amino acids form a pair.
Most read news
Other news from the department science

Get the life science industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.
Most read news
More news from our other portals
Last viewed contents
Category:Osmotic_diuretics
Efficiently producing fatty acids and biofuels from glucose






















































