Snapshots of the flu virus replication machine in action
Researchers from EMBL Grenoble have, for the first time, observed different functional states of the influenza virus polymerase as it is actively transcribing. These results, published in Nature Structural & Molecular Biology, provide valuable information for the next generation of anti-influenza drugs.
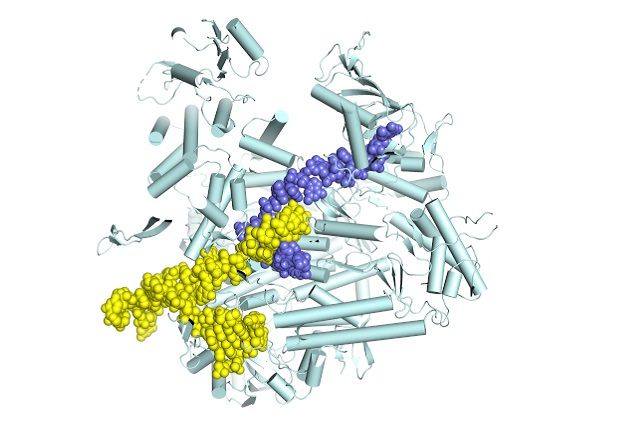
Influenza polymerase (light cyan) in the process of synthesising messenger RNA (slate blue) by copying the genomic viral RNA template (yellow).
Stephen Cusack / EMBL
When a virus infects and enters a host cell, the genomic material in the virus is both replicated to produce multiple copies of itself and transcribed into viral messenger RNA (mRNA). The viral mRNA can be read by the host cell’s protein production machinery, tricking it into making viral proteins. The viral proteins package the copies of the viral genome to make progeny viruses that are released from the cell to infect new hosts. An enzyme called a polymerase is responsible for both the transcription and replication of the viral genome and is therefore crucial to the successful propagation of the virus. The influenza virus, with genomic material made of RNA rather than DNA, is no exception to this modus operandi.
“Studying and understanding the unique mechanisms of transcription and replication used by influenza virus is essential to fight its spread,” explains EMBL group leader Stephen Cusack.
A decade long study
Stephen Cusack and his research group at EMBL Grenoble started to work on influenza polymerase more than 20 years ago. In 2014, the group published the first crystal structures of the complete polymerase machine. However, attempts to structurally characterise the different states of actively transcribing influenza polymerase have so far been unsuccessful.
“All studies so far have looked at the resting structure of the polymerase machine; we’ve never observed it actually doing anything,” says Tomas Kouba, a postdoc in the group who carried out much of the work.
Using X-ray crystallography and cryo-electron microscopy, both performed on state-of-the-art equipment at the European Synchrotron Radiation Facility in Grenoble, the group were able to determine, for the first time, atomic-resolution structures of different functional states of the influenza polymerase as it is actively transcribing genomic RNA into mRNA. In particular, it provides the first characterisation of the movements of the polymerase during the so-called nucleotide addition cycle, whereby each successive nucleotide is added to the growing mRNA chain.
Target point for new drugs
Despite its familiarity, the influenza virus – some strains of which are among the top ten most dangerous viruses in the world – is far from being well understood. Up to 500,000 people worldwide die from influenza each year, according to the World Health Organization, with potentially much higher mortality rates when a new pandemic flu strain emerges. Vaccination is not always effective and anti-influenza drugs are needed as a complementary treatment option.
As the viral polymerase is essential for the replication of the flu virus, it is a prime target for the development of new anti-influenza drugs. As such, the results gained by the EMBL group give new insights to help design the next generation of anti-influenza drugs. The advantage of drugs that stop the polymerase functioning is that it is much less likely that the virus will mutate in a way that would render the drug useless.
“Our results contribute the first snapshots of a movie that will show the complete transcription cycle of influenza polymerase from initiation to termination,” explains Cusack. The group will continue to use the powerful technique of cryo-electron microscopy to fill in the missing steps.