Bioinformatics platform for the genome-based taxonomical classification of bacteria and archaea
Researchers develop Type Strain Genome Server (TYGS)
"TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy", is the title of the paper published in the current issue of the internationally journal Nature Communications. In this paper, Dr. Jan P. Meier-Kolthoff and Ass. Prof. Dr. Markus Göker, both of the Department for bioinformatics at the Leibniz Institute DSMZ-German Collection of microorganisms and Cell Cultures, share their findings regarding the new Type Strain Genome Server TYGS.
Leibniz-Institut DSMZ
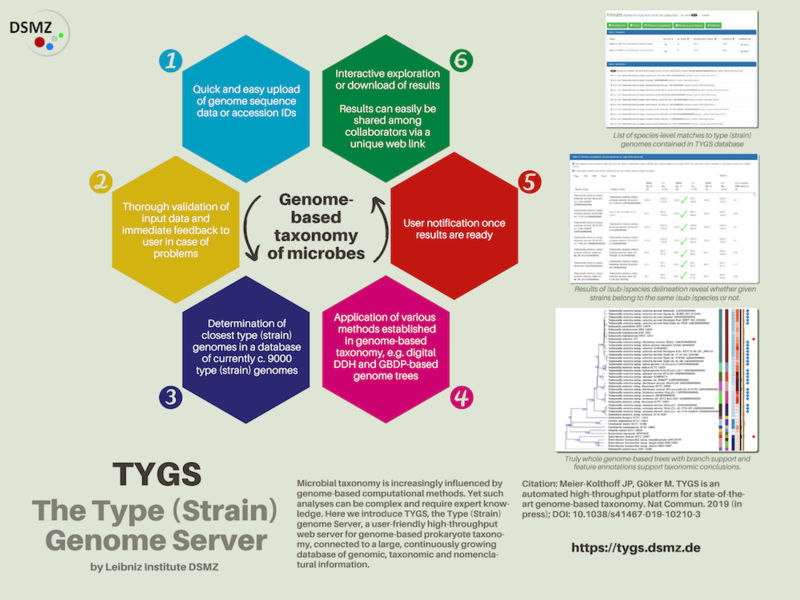
Scientists at the DSMZ developed this Type (Strain) Genome Server as a bioinformatics platform for the genome-based taxonomy of bacteria and archaea. The server is based on a database with weekly updates, which links comprehensive information on taxonomy and nomenclature to almost 9,000 microbial type strain genome sequences and essentially all type strain 16S sequences.
The Type (Strain) Genome Server uses a high-throughput process to automatically determine the type strain genome sequences most closely related to one (or up to several) user-defined genome sequences, and from there calculates, amongst others, a true genome-based family tree, a categorization of species and subspecies using digital DNA:DNA hybridization, and all differences in genomic GC-Content. For this, the TYGS uses a number of scientifically established and frequently cited methods including the Genome-to-Genome Distance Calculator (GGDC) or a phylogenomic process called Genome BLAST Distance Phylogeny (GBDP).
The user receives a specific link to an interactive website of the TYGS-server containing all relevant information in form of tables, modifiable figures ready for publication, references on taxonomy literature as well as complete texts on methods and results. The data can be integrated directly into microbiological studies and papers; in addition, the user is also able to exchange them with cooperation partners via the web link provided. Should the phylogenetic environment be thought to not yet contain genome-sequenced type strains, the user may conveniently use TYGS on the basis of this result to request a more comprehensive tree analysis of the 16S rRNA gene sequences.
Original publication
Other news from the department science

Get the life science industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.