Unlimited access to microbiological research data
Update of the freely accessible database BacDive enables access to more than 900,000 metadata
The scientists Dr. Lorenz Reimer, Joaquim Sardà and Prof. Dr. Jörg Overmann from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures have extended the database BacDive extensively with the update in April 2019. Since 2012, the bioinformaticians of the DSMZ have been developing BacDive (the Bacterial Diversity Metadatabase), a database that is unique in the world and makes data on bacteria and archaea that are not yet available freely accessible. The development follows the FAIR principles, according to which scientific data should be findable, accessible, interoperable and re-usable.
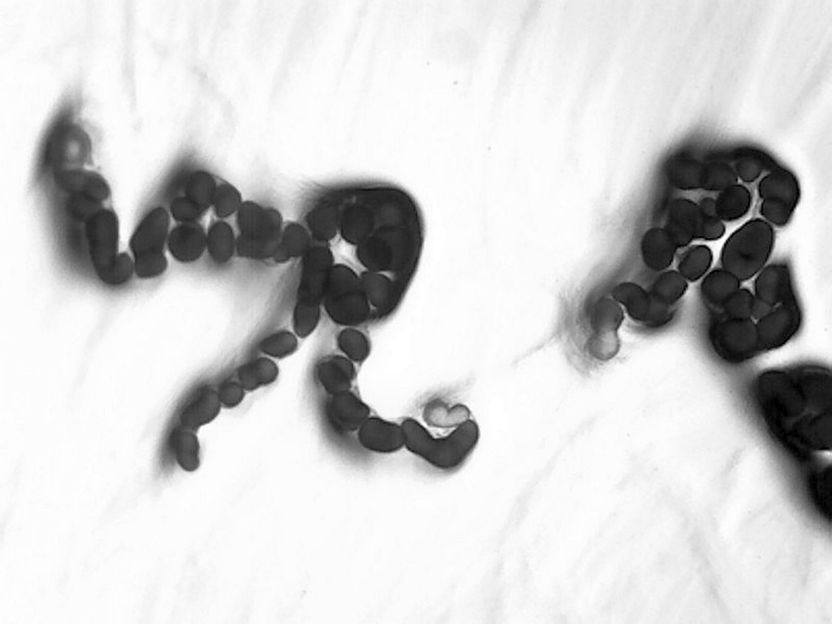
Microscopic image of the bacterium Cystobacter ferrugineus (DSM 14716)
DSMZ
More than 600 data fields available
The possibilities of using BacDive are continuously being expanded, currently scientists can use more than 600 data fields to search for microbiological information. The repertoire includes initial species descriptions and metabolic profiles as well as data on enzymatic activities and antibiotic resistance. In addition, BacDive offers 9,000 Analytical Profile Tests (API) for over 5,000 bacterial strains, the largest publicly available API data collection worldwide.
External Data Integration and Interoperability
In order to provide the scientific community with a comprehensive database, the DSMZ scientists also continuously maintain the microbiological metadata of other European collections in BacDive. With the current update, DSMZ integrated data from more than 19,505 bacterial strains from the Swedish Culture Collection University of Gothenburg (CCUG). BacDive thus provides up-to-date information on 80,584 bacteria and archaea. For scientific work with cross-source data sets, the various information sources must be linked as directly as possible via unique identifiers. For example, direct links to the 16S rRNA sequence of a bacterial strain in the database SILVA or to the enzymes involved in metabolic reactions in the database BRENDA have been established. Via links in PubMed, NCBI Taxonomy, NCBI Nucleotide, but also in species descriptions of Wikipedia, users can access the structured metadata in BacDive.
Organizations
Related link
Other news from the department science

Get the life science industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.
Most read news
More news from our other portals
Last viewed contents
Genedata Begins High-Throughput Screening Collaboration with NIH Chemical Genomics Center



















































