A simple solution to a complex problem
Researchers use a novel approach to identify a transport protein in mycobacteria
A team headed by Dr. Claudia Jessen-Trefzer of the University of Freiburg’s Institute for Pharmaceuticals Sciences has for the first time identified a transport protein in mycobacteria which is responsible for the uptake of the nutrient L-arabinofuranose. The lead authors of the study, Miaomiao Li of the Institute for Pharmaceuticals Sciences, Christoph Müller of the Institute for Biochemistry and Klemens Fröhlich of the Institute of Molecular Medicine and Cell Research at the University of Freiburg, used a novel approach which could simplify the identification of transport proteins in mycobacteria in the future. This class of proteins could play a key role in the development of new types of medications to tackle mycobacteria and treat diseases like tuberculosis in humans.
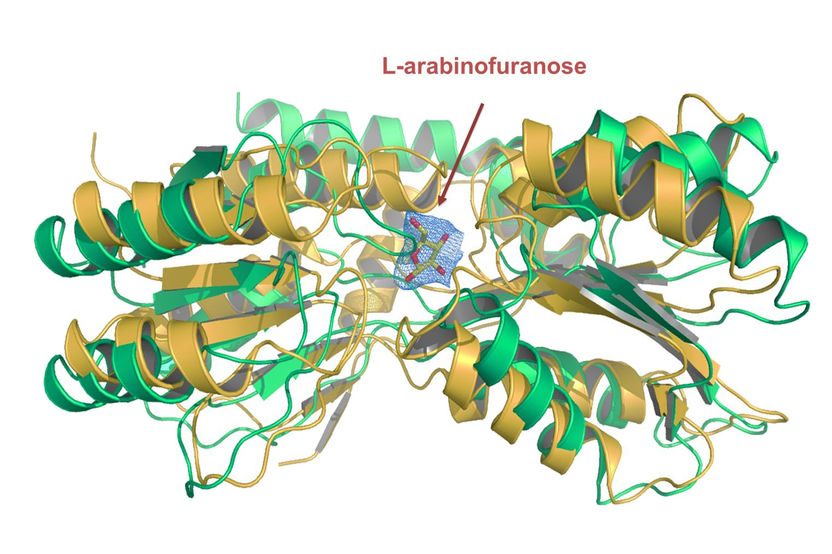
The crystal structure of the transport protein’s substrate binding domain, which is responsible for absorbing the nutrient L-arabinofuranose.
Claudia Jessen-Trefzer
Transport proteins are located in the membrane of the cell and are responsible for absorbing nutrients into the cell and expelling toxic substances out of it. They therefore drive processes which are vital to the cell’s survival. Until now, little was known about the proteins because their chemical properties make them difficult to examine. The Freiburg researchers characterized the protein on the basis of its substrate, the nutrient L-arabinose, with which the protein bonds. A subgroup of the bacterium, Mycobacterium smegmatis, served as the model organism.
The team developed a method in which they carried out gene expression - the targeted expression of genes and the translation into proteins - via the substrate. Then they investigated the proteins isolated from the membrane using mass spectrometry. This provided the Freiburg researchers with a relatively simple way of identifying the transport proteins which are responsible for absorbing a certain nutrient. In addition, they were able to identify the L-arabinofuranose transporter whose characteristics could be determined with protein crystallography. The team managed to vastly simplify the identification of transport proteins in mycobacteria, thereby contributing to the search for potential target proteins for antibiotics development.
Original publication
Miaomiao Li; Christoph Müller; Klemens Fröhlich; Oliver Gorka; Lin Zhang; Olaf Groß; Oliver Schilling; Oliver Einsle; Claudia Jessen-Trefzer; "Detection and characterization of a mycobacterial L-arabinofuranose ABC-transporter identified with a rapid lipoproteomics protocol"; Cell Chemical Biology; 2019.
Other news from the department science
Most read news
More news from our other portals
See the theme worlds for related content
Topic World Mass Spectrometry
Mass spectrometry enables us to detect and identify molecules and reveal their structure. Whether in chemistry, biochemistry or forensics - mass spectrometry opens up unexpected insights into the composition of our world. Immerse yourself in the fascinating world of mass spectrometry!
Topic World Mass Spectrometry
Mass spectrometry enables us to detect and identify molecules and reveal their structure. Whether in chemistry, biochemistry or forensics - mass spectrometry opens up unexpected insights into the composition of our world. Immerse yourself in the fascinating world of mass spectrometry!





















































