How bacteria turn off an antibiotic
Advertisement
Many common antibiotics are increasingly losing their effectiveness against multi-resistant pathogens, which are becoming ever more prevalent. Bacteria use natural means to acquire mechanisms that protect them from harmful substances. For instance against the agent albicidin: Harmful gram-negative bacteria possess a protein that binds and inactivates albicidin. The underlying resistance mechanism has been investigated at atomic resolution by scientists from the Helmholtz Centre for Infection Research (HZI) and the associated Helmholtz Institute for Pharmaceutical Research Saarland (HIPS). The scientists discovered that this protein not only binds the antibiotic, but also promotes its chemical modification. Concurrently, it promotes its own production such that it can inactivate albicidin even more efficiently.
The Gram-negative Klebsiella pneumoniae bacterium often becomes resistant to common antibiotics.
NIAID/CC BY 2.0
Pathogenic bacteria have developed various mechanisms protecting them from antibiotics. Some simply expel the substances that are toxic to them, whereas others alter their cell wall structure so that antibiotics never manage to get into the cell. Other pathogens change the target structure attacked by antibiotics – or simply modify the antibiotic itself. A different trick is used by bacteria of the Klebsiella genus: They possess a protein called AlbA that binds to albicidin and protects the bacteria from the effect of this antibiotic. The underlying defence mechanism has been investigated by scientists from the Helmholtz Centre for Infection Research (HZI), the Helmholtz Institute for Pharmaceutical Research Saarland (HIPS) – which is a facility of the HZI run in cooperation with the Saarland University (UdS) – and the German Center for Infection Research (DZIF).
The pathogen Klebsiella pneumoniae is the most common cause of bacterial sepsis and hospital-acquired inflammations of the lung, but can also elicit urinary tract infections. Klebsiella bacteria belong to the group of Gram-negative bacteria that are difficult to treat and often develop resistance to multiple antibiotics. HZI scientists have already discovered an effective new class of agents against Gram-negative pathogens: the cystobactamids. "This substance class is similar to albicidin, to which many Klebsiella strains are already resistant," says Junior Prof Jesko Köhnke, who directs the "Structural Biology of Biosynthetic Enzymes" young investigator research group at the HIPS. "We wanted to know if the AlbA protein is effective against cystobactamids as well, so we analysed the mechanism of resistance on a molecular level."
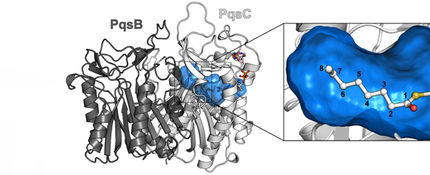
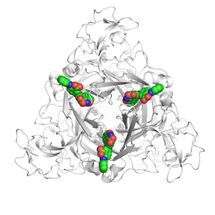
To this end, an interdisciplinary team of scientists around Jesko Köhnke came together, in which the department of Prof Mark Brönstrup took on the synthesis, the research group of Prof Olga Kalinina contributed the bioinformatics, and the department of Prof Rolf Müller was responsible for the activity tests and gene expression profiles. Initially, the three-dimensional structure of the AlbA protein with the antibiotic albicidin bound to it was elucidated by X-ray diffraction analysis. The scientists noticed that, in the course of evolution, a small binding pocket for the antibiotic had become an elongated tunnel perfectly accommodating albicidin. But the structural analysis had another surprising result: The bound albicidin was chemically modified. "Originally, we presumed that AlbA would simply take up albicidin – much like a sponge takes up water – and thereby eliminate it transiently. But then we found that it actually modifies albicidin," Köhnke says. Further experiments have shown that the activity of the modified albicidin is lower, i.e. it is less harmful to the bacteria.
The scientists discovered that AlbA possesses not only the binding site for the antibiotic, but also another one for DNA. This section allows it to bind to the genetic material of the bacterial cell – and to do so exactly at the site at which the production of AlbA is controlled. In laboratory experiments with Klebsiella pneumoniae, the team of Jesko Köhnke was able to show that the administration of albicidin causes the gene bearing the blueprint for AlbA to be read 3000 times more often after four hours than in the absence of the antibiotic. "The mechanism used by Klebsiella against albicidin is extremely effective: As soon as they are exposed to it they scavenge the first molecules of the agent with their AlbA protein. At the same time, they produce increasing amounts of AlbA. But we were also able to show that these mechanisms are not sufficient for resistance yet, which is what we want to investigate in more detail in the future," Köhnke says.
Comparative experiments have shown that the related cystobactamid agent is bound markedly less strongly by AlbA. In addition, the production of AlbA is not stimulated such that the Klebsiella bacteria have not been able to develop resistance to cystobactamid with this mechanism. "Yet, the bacteria are resistant to cystobactamid. We therefore want to elucidate the underlying mechanisms at the greatest possible detail in order to be able to develop agents that circumvent these resistances," Köhnke says.