Gene recombination deactivates retroviruses during invasion of host genomes
Advertisement
Most vertebrate genomes contain a surprisingly large number of viral gene sequences – about eight percent in humans. And yet how do exogenous viruses – apparently having invaded from outside – manage to become integrated into the host genome? Answers to this question are provided in a study by an international team of researchers led by Alex Greenwood of the Leibniz Institute for Zoo and Wildlife Research (Leibniz-IZW) in Berlin.

Holgi, pixabay.com, CC0
Working with the example of koalas, the researchers have now identified key stages in the process, called “endogenization”, by which a host is invaded by exogenous retroviruses. The scientists also uncovered a process by which the host genome mounts a defense against the invaders.
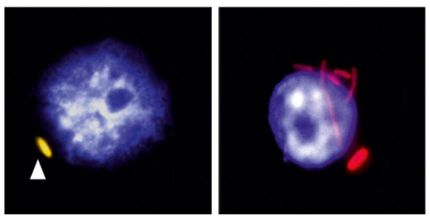
By using the enzyme reverse transcriptase, retroviruses convert their RNA genomes into DNA in order to integrate retroviral genes into the genome of the host. If the retroviruses manage to infect germ cells, the infected host will then transmit the viral sequences to its offspring. Sequences of human endogenous retroviruses, referred to as HERVs, comprise around eight percent of the human genome. Although recombination, and later mutations, led to the inactivation of most HERVs in the distant past, some viral sequences are still being read and encode for proteins. They are thought to trigger cancer and autoimmune diseases in some cases.
As was the case with all mammals and birds, our ancestors were infected by these viruses millions of years ago. “The original viruses have long become extinct, and the sequences integrated into the host germ line have been altered so considerably by mutation that it is hard to determine which changes were important and which processes were initially involved,” explained Alex Greenwood, head of the Department of Wildlife Diseases at the Leibniz-IZW and senior author of the study. “With koalas, however, we are able to follow this process live, as it were, since the retrovirus is currently transitioning into the koala genome and the process has not yet been completed.”
The sequencing of the koala genome (published recently in Nature Genetics), also conducted with the involvement of the Leibniz-IZW scientists, featured the use of a high-throughput sequencing method known as long-read sequencing. This method enabled the researchers to identify not only repetitive DNA regions such as retroviruses, but also to identify flanking sequences from the host genome, and to explore the process of their invasion using the example of this marsupial. For their latest study, the team examined DNA samples from 169 wild koalas, as well as from two zoo koalas and six historic museum specimens.
The koala genome project and other previous studies had discovered that the quantity of endogenous koala retroviruses (KoRV) differs greatly by region, which can be explained by the natural barriers along the east coast of Australia. These barriers prevent the mixing of populations, and therefore also unchecked proliferation of the virus: whereas in the northeast in Queensland all animals are already infected, the researchers found that the further south they went, the lower the number of KoRVs per koala.
Even in koalas from less infected populations, the scientists found highly modified viral sequences (recKoRV) in the koala genome that differed from the intact virus. These sequences appear to be the result of recombination with very old viral genomic elements also present in other Australian marsupials.
What are the implications of these findings? “We believe that the first ancient viral components – that became fixed in the koala genome and are no longer pathogenic – defend the host genetic material: by recombining, they incapacitate the new viral sequences, even if the older viruses now have only little resemblance to their original sequences,” stated Alex Greenwood. “That’s good news for the koalas! Because recombinant virus sequences are likely to be less harmful than the original.”
In addition, 17 independent recombination events between KoRV and an old, degraded virus fragment (PhER) must have taken place – exclusively between KoRV and PhER. One of these recombinants (recKoRV1) is very common in koalas, with several copies existing in each animal. This suggests that recKoRV1 was independently created several times at different points in the past.
“Regardless of which region of Australia the sample came from, some sequences of endogenous KoRV were always recombinant and highly degraded. We assume that this process represents a very early stage in the endogenization of exogenous retroviruses,” stated Alex Greenwood.
Original publication
Löber U, Hobbs M, Dayaram A, Tsangaras K, Jones K, Alquezar-Planas DE, Ishida Y, Meers J, Mayer J, Quedenau C, Chen W, Johnson RN, Timms P, Young P, Roca AL, Greenwood AD; "Degradation and remobilization of endogenous retroviruses by recombination during the earliest stages of a germ line invasion"; PNAS; 2018.
Johnson RN et al.; "Adaptation and conservation insights from the koala genome"; Nature Genetics; 2018; 50, 1102–1111.