DNA enzyme shuffles cell membranes a thousand times faster than its natural counterpart
A new synthetic enzyme, crafted from DNA rather than protein, flips lipid molecules within the cell membrane, triggering a signal pathway that could be harnessed to induce cell death in cancer cells.
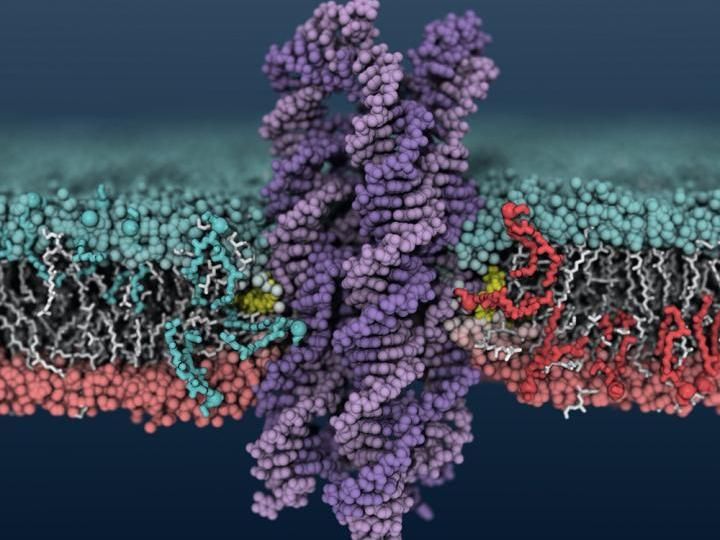
A synthetic DNA enzyme inserts into a cell membrane, causing lipids to shuffle between the inner and outer membrane layers.
Image courtesy of Christopher Maffeo
Researchers at University of Illinois at Urbana-Champaign and the University of Cambridge say their lipid-scrambling DNA enzyme is the first in its class to outperform naturally occurring enzymes - and does so by three orders of magnitude.
"Cell membranes are lined with a different set of molecules on the inside and outside, and cells devote a lot of resources to maintaining this," said study leader Aleksei Aksimentiev, a professor of physics at Illinois. "But at some points in a cell's life, the asymmetry has to be dismantled. Then the markers that were inside become outside, which sends signals for certain processes, such as cell death. There are enzymes in nature that do that called scramblases. However, in some diseases where scramblases are deficient, this doesn't happen correctly. Our synthetic scramblase could be an avenue for therapeutics."
Aksimentiev's group came upon DNA's scramblase activity when looking at DNA structures that form pores and channels in cell membranes. They used the Blue Waters supercomputer at the National Center for Supercomputing Applications at Illinois to model the systems at the atomic level. They saw that when certain DNA structures insert into the membrane - in this case, a bundle of eight strands of DNA with cholesterol at the ends of two of the strands - lipids in the membrane around the DNA begin to shuffle between the inner and outer membrane layers.
To verify the scramblase activity predicted by the computer models, Aksimentiev's group at Illinois partnered with professor Ulrich Keyser's group at Cambridge. The Cambridge group synthesized the DNA enzyme and tested it in model membrane bubbles, called vesicles, and then in human breast cancer cells.
"The results show very conclusively that our DNA nanostructure indeed facilitates rapid lipid scrambling," said Alexander Ohmann, a graduate student at Cambridge and a co-first author of the paper along with Illinois graduate student Chen-Yu Li. "Most interestingly, the high flipping rate indicated by the molecular dynamics simulations seems to be of the same order of magnitude in experiments: up to a thousand-fold faster than what has previously been shown for natural scramblases."
On its own, the DNA scramblase produces cell death indiscriminately, Aksimentiev said. The next step is to couple it with targeting systems that specifically seek out certain cell types, a number of which have already been developed for other DNA agents.
"We are also working to make these scramblase structures activated by light or some other stimulus, so they can be activated only on demand and can be turned off," Aksimentiev said.
"Although we have still a long way to go, this work highlights the enormous potential of synthetic DNA nanostructures with possible applications for personalized drugs and therapeutics for a variety of health conditions in the future," Ohmann said.