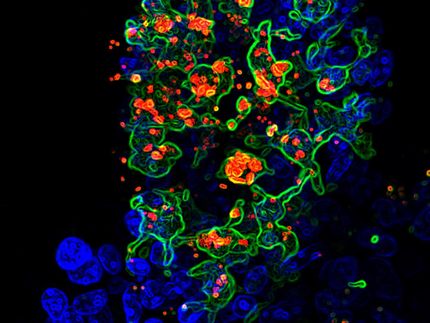
A factor important for ZIKA Virus host species restriction uncovered
Advertisement
Princeton University researchers Qiang Ding, Alexander Ploss, and colleagues have identified one of the mechanisms by which Zika virus (ZIKV) circumvents immune control to replicate in human cells.
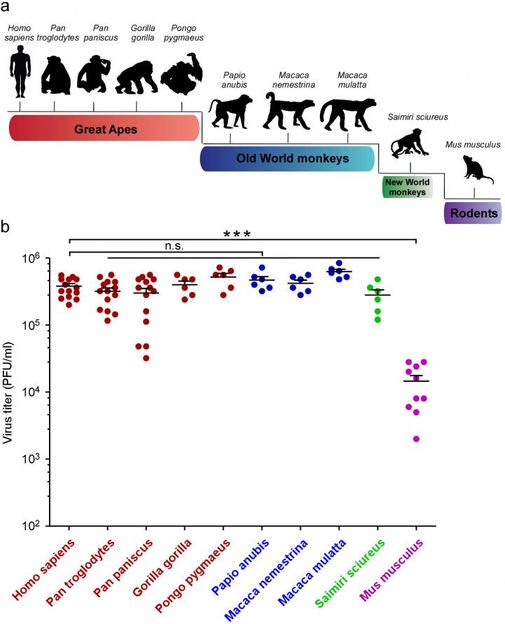
Top: A schematic representation of the evolutionary relationship between mammal species that were tested for ZIKA susceptibility. Bottom: Princeton biologists Qiang Ding and Alexander Ploss analyzed dermal fibroblasts (skin cells) isolated from the different species and infected with the ZIKA virus. Viral titer was determined three days after infection.
Qiang Ding and Alexander Ploss, Princeton University
In 2013, and again in 2015, the world witnessed devastating outbreaks of ZIKV, a hitherto obscure member of the flavivirus family. Although infection with this virus usually produces no symptoms in most adult humans, it can cause severe pre-natal neurodevelopmental disorders. It also sometimes sparks a serious autoimmune disorder, and in rare cases causes male sterility in adults. It's clear the virus poses a major threat to human health, but people are not its native hosts. ZIKV can spread between people via sexual contact, but the virus likely dwells in some wild animal population - probably non-human primates - before being picked up by mosquitos and transmitted to humans. In fact, ZIKV was first detected in a monkey in Africa in 1947. However, the identity of the virus's reservoir host is still unknown, and no one has yet conducted any comprehensive examination of which species may host ZIKV. This was the problem the Princeton scientists set out to address.
"We systematically tested the ability of ZIKV to infect cells from humans, great apes, New World and Old World monkeys, and mice," explains Ploss, an assistant professor in the Department of Molecular Biology.
Dermal fibroblasts (skin cells) from these animals were exposed to ZIKV, and the level of viral replication was monitored. Mice are known to be resistant to the virus, and as expected, the mouse cells repelled viral invasion. However, the cells of every primate species tested, from humans to squirrel monkeys, were susceptible to ZIKV and produced large amounts of virus.
What accounts for the failure of ZIKV to mount infections of mouse cells? Prior studies have shown that proteins called interferons are essential for this protection; mice that have been genetically engineered to be deficient in interferon alpha/beta; responses are susceptible to ZIKV infection. Interferons are secreted proteins that are critical for stimulating inflammation and triggering immune system defenses against viruses, bacteria, parasites and cancers. Virally infected cells recognize the presence of viruses using one of several different immune "pattern receptors" that bind to pathogen DNA or RNA and respond by stimulating interferon secretion.
To determine which immune pattern receptor is important for recognizing ZIKV infection, Ding and colleagues examined whether exposure to ZIKV impairs the function of any these pathways in human cells. They found that ZIKV shuts down signaling by the cGAS-STING DNA sensing pathway. When the researchers examined the components of this pathway, they observed that the STING protein was cleaved into two fragments in ZIKV-infected cells. This is consistent with earlier studies, which had shown that a ZIKV protein called NS2B3 also targets this pathway by destroying cGAS.
Ding et al. discovered that NS2B3 cleaves human STING, and identified the site at which this cleavage takes place. The mouse version of STING has different amino acids at the cleavage site than does the human protein, so NS2B3 cannot cleave mouse STING.
"Notably," says Ploss, "this cleavage site appears to also be shared by NS2B3 of other flaviviruses including West Nile virus, Japanese encephalitis virus, and dengue virus, but not yellow fever virus."
Interestingly, the STING proteins of several primates, including chimpanzees, rhesus macaques, and squirrel monkeys, also differ from human STING at the NS2B3 cleavage site, and STING from these species was resistant to cleavage by NS2B3. Even when the human amino acids were swapped into the primate proteins at the cleavage site, only chimpanzee STING became vulnerable to NS2B3 cleavage. This suggests that chimpanzee STING may share some structural feature or other characteristic with human STING that more distantly related animals do not. Nonetheless, ZIKV replicates in all these species, which suggests the virus employs other mechanisms besides STING degradation to evade primate immune defenses; some of these have already been identified.
To test whether mouse STING's resistance to cleavage by NS2B3 accounts for the animal's ability to curb ZIKV, Ding et al. used CRISPR/CAS9 technology to remove STING from mouse cells. This change rendered the cells susceptible to ZIKV infection in vitro. But even though the virus could replicate in these cultured mouse cells, the authors found that STING-deficient mice were still able to resist ZIKV infection, and did not become ill. This indicates that there exist other mechanisms that help keep the virus in check in mice.
As Ploss points out: "Our data point to a molecular basis for the restricted host tropism of ZIKV. This could aid in the development of mice with inheritable susceptibility to ZIKV and potentially other flaviviruses. Such a model would pave the path to testing novel drug and vaccine candidates and for gaining a deeper understanding of how ZIKV and other viruses cause disease."