Researchers find algorithm for large-scale brain simulations on next-generation supercomputers
Ready for Exascale
The human brain is an organ of incredible complexity, composed of a hundred billion interconnected nerve cells. However, even with the help of the most powerful supercomputers available, it is currently impossible to simulate the exchange of neuronal signals in networks of this size. Researchers of Forschungszentrum Jülich, Germany, RIKEN, Kobe and Wako, Japan, and the KTH Royal Institute of Technology, Stockholm Sweden, have made a decisive step towards creating the technology to achieve simulations of brain-scale networks on future supercomputers of the exascale class. Simultaneously, the new algorithm significantly speeds up brain simulations on existing supercomputers.
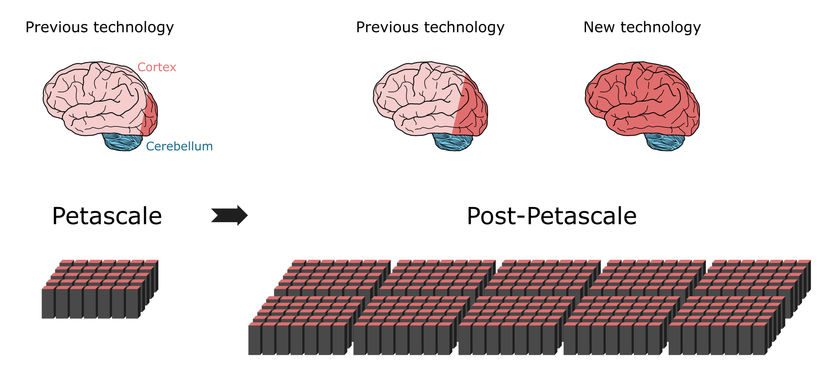
Previous simulation technology can represent about 1% of nerve cells (neurons) in a human brain (dark red area of brain on left) using a petascale supercomputer (bottom left). Only small progress (dark red area of center brain) would be possible on next generation of supercomputers that will exceed the performance of today's high-end supercomputers by a ten- to a hundredfold. Larger parts of the human brain can be represented with the new technology, using the same amount of computer memory (bottom right) 10% of the brain are approximately the size of the whole cortex (dark red area of brain on right), which is essential for higher processing. A large part of the other neurons are in the cerebellum (blue).
Copyright: Forschungszentrum Jülich
"Since 2014, our software can simulate about one percent of the neurons in the human brain with all their connections", says Markus Diesmann, Director at the Jülich Institute of Neuroscience and Medicine (INM-6). In order to achieve this impressive feat, the software requires the entire main memory of petascale supercomputers, such as the K computer in Kobe and JUQUEEN in Jülich. Diesmann has been working for more than twenty years on the simulation software NEST - a free, open-source simulation code in widespread use by the neuroscientific community and a core simulator of the European Human Brain Project. With NEST, the behavior of each neuron in the network is represented by a handful of mathematical equations. In HBP, Diesmann leads projects in the areas of Theoretical Neuroscience and on the High-Performance Analytics and Computing Platform. Future exascale computers, such as the post-K computer planned in Kobe and JUWELS in Jülich, will exceed the performance of today's high-end supercomputers by a ten- to a hundredfold. For the first time, researchers will have the compute power available to simulate neuronal networks on the scale of the human brain.
Seemingly a dead end
While current simulation technology enabled researchers to begin studying large neuronal networks, it also represented a dead end on the way to exascale technology. Supercomputers are composed of about a hundred thousand small computers, called nodes, each equipped with a number of processors doing the actual calculations. "Before a neuronal network simulation can take place, neurons and their connections need to be created virtually, which means that they need to be instantiated in the memory of the nodes. During the simulation a neuron does not know on which of the nodes it has target neurons, therefore, its short electric pulses, need to be sent to all nodes. Each node then checks which of all these electric pulses are relevant for the virtual neurons that exist on this node", says Susanne Kunkel of KTH Royal Institute of Technology in Stockholm.
The current algorithm for network creation is efficient because all nodes construct their particular part of the network at the same time. However sending all electric pulses to all nodes is not suitable for simulations on exascale systems. "Checking the relevance of each electric pulse efficiently requires one Bit of information per processor for every neuron in the whole network. For a network of 1 billion neurons a large part of the memory in each node is consumed by just this single Bit of information per neuron", adds Markus Diesmann.
This is the main problem when simulating even larger networks: the amount of computer memory required per processor for the extra Bits per neuron increases with the size of the neuronal network. At the scale of the human brain, this would require the memory available to each processor to be one hundred times larger than in today’s supercomputers. This, however, is unlikely to be the case in the next generation of supercomputers. The number of processors per compute node will increase but the memory per processor and the number of compute nodes will rather stay the same.
Breakthrough by new algorithm
The breakthrough reported in a recent publication is a new way of constructing the neuronal network in the supercomputer. Due to the algorithms, the memory required on each node no longer increases with network size. At the beginning of the simulation, the new technology allows the nodes to exchange information about who needs to send neuronal activity data to whom. Once this knowledge is available, the exchange of neuronal activity data between nodes can be organized such that a node only receives the information it requires. An additional Bit for each neuron in the network is no longer necessary.
A beneficial side effect
While testing their new ideas, the scientists made an additional key insight, reports Susanne Kunkel: "When analyzing the new algorithms we realized that our novel technology would not only enable simulations on exascale systems but it would also make simulations faster on presently available supercomputers.” In fact, as the memory consumption is now under control, the speed of simulations becomes the main focus of further technological developments. For example, a large simulation of 0.52 billion neurons connected by 5.8 trillion synapses running on the supercomputer JUQUEEN in Jülich previously required 28.5 minutes to compute one second of biological time. With the improved data structures simulation time is reduced to 5.2 minutes.
"With the new technology we can exploit the increased parallelism of modern microprocessors a lot better than previously, which will become even more important in exascale computers” remarks Jakob Jordan, lead author of the study, at Forschungszentrum Jülich. “The combination of exascale hardware and appropriate software brings investigations of fundamental aspects of brain function, like plasticity and learning unfolding over minutes of biological time within our reach" adds Markus Diesmann.
With one of the next releases of the simulation software NEST, the researchers will make their achievement freely available to the community as open source.
"We have been using NEST for simulating the complex dynamics of the basal ganglia circuits in health and Parkinson’s disease on the K computer. We are excited to hear the news about the new generation of NEST, which will allow us to run whole-brain-scale simulations on the post-K computer to clarify the neural mechanisms of motor control and mental functions" says Kenji Doya of Okinawa Institute of Science and Technology (OIST).
"The study is a wonderful example of the international collaboration in the endeavor to construct exascale computers. It is important that we have applications ready that can use these precious machines from the first day they are available" concludes Mitsuhisa Sato of the RIKEN Advanced Institute for Computer Science in Kobe.